What is CSV?
A comma-separated values (CSV) file stores tabular data (numbers and text) in plain text. Plain text means that the file is interpreted a sequence of characters, so that it is human-readable with a standard text editor. Each line of the file is a data record. Each record consists of one or more fields, separated by commas. The use of the comma as a field separator is the source of the name for this file format.
See http://en.wikipedia.org/wiki/Comma-separated_values
You can use tabulator ('\t'), semicolon (';') or comma (',') as a delimiter.
Example Files
In the github repository, you can find some CSV Example Files.
Import CSV
To start the CSV Import, right click the project in the Import Browser you want to upload data to and select Import Data -> Import CSV.
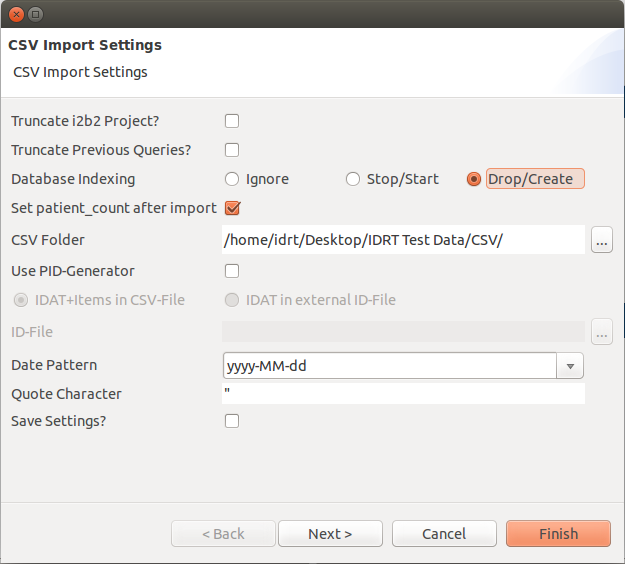
A new window opens, where you can set different options.
Name | Description |
|---|---|
Truncate i2b2 Project? | If you check this, the project will be truncated before the data is uploaded. |
Truncate Previous Queries? | This will truncate the previous queries tables. |
Database Indexing | Ignore: Don't touch the indexes; |
Set patient_count after import | This will fill the c_totalnum column. |
CSV Folder | Here you can select the folder that contains all CSV files you want to upload. |
Use PID Generator | This will use the PID Generator, which has to be configured separately (see Options). |
IDAT Options | Select whether your identifying data is in the same or in an extra file. |
ID-File | Select the identifying data file here. |
Date Pattern | Here you can select the date pattern that your data uses (e.g. dd-MM-yyyy). |
Quote Character | Here you can set the quote character that you use in your CSV files. |
Save Settings? | This will save the settings for the next import. Note: The truncate checks are never saved. |
Set your variables and continue to page 3.
On the left hand side you can see all CSV files in the selected directory. A green check mark lets you know, that a config file has been found. If there is currently no config file present, a red X is shown. On the right hand side the configuration for the selected CSV file is shown. The table consists of five columns:
Item: The column name in the CSV file.
Name: The name, the item should have in i2b2. You can e.g. rename cryptic column names.
Tooltip: The tooltip the item should have in i2b2. You can set a tooltip for better understanding, or the unit of an item for example.
Datatype: The datatype the item has. You can select either Integer, Float, Date, or String.
Metadata: This tells the Import Browser, where to look for specific items, such as the id of the patient, the import date, start date and so on. You can also ignore items. These will not end up in the i2b2 database.
The metadata item PatientID/ObjectID has to be present in each configuration file. Without it, the Import Browser won't let you continue and displays an error.
The ObjectID is the same as the PatientID, but the file is imported using modifiers instead. This way more than one row per patient is allowed. The Import Browser will tell you, if you used the Guess Schema functionality whether you should use the ObjectID instead.
The next table describes the buttons in the CSV Import Settings page.
Name of the Button | Description |
|---|---|
Headline | If your CSV file does not start directly at the beginning, you can set the starting row here. |
Clear Table | This will delete your configuration for the selected file. |
Guess Schema | You can let the Import Browser try to guess the schema of the file. You can set in the Options how many rows the Import Browser should look through. |
Guess All Schemata | This will guess the schema for all files in the directory. |
If you hit Finish the upload starts. You can observe the progress in the progress bar of the Import Browser.


3 Comments
Anonymous
Anonymous
Wait, what? Observations that are not modifiers can have more than one row per patient. In fact, a patient usually has multiple diagnoses, meds, orders, etc. recorded during each visit. Modifier as used by i2b2 means contextual info about a specific observation.
These docs would be easier to understand if the same jargon was used as is used in i2b2's documentation, at least that way we only have one jargon to learn.
Anonymous
Do the example files contain someplace the standard demographic fields used by PATIENT_DIMENSION or do they rely on existing entries in PATIENT_DIMENSION?
What happens if the source data contains a duplicate of something that already exists in PATIENT_DIMENSION, CONCEPT_DIMENSION, MODIFIER_DIMENSION, or OBSERVATION_FACT?