Page History
| Info Box | ||
|---|---|---|
| ||
Downloads:
|
...
Documentation:
|
| Warning | ||
|---|---|---|
| ||
Note that we have discovered an issue loading the CRC Stored Procedures on SQL Server and have found a workaround. Please be sure to read the updated content of Step 3.4.4 Create Crcdata Stored Procedures before performing this step. |
1.7.13 Release Notes
i2b2 1.7.13 offers support for SAML federated login, enhanced security due to improvements found via an internal Veracode scan, a client-based user registration tool, support for Synthea synthetic data for testing, and a variety of other bugfixes and performance improvements. i2b2 1.7.13 has been tested with SHRINE 3.2.1.
| UI Button | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| Table of Contents |
|---|
Highlight of Features
...
Top New Features
LOTS OF TABS!
| Description | |
|---|---|
| SAML Authentication | |
| User Account Registration Tool | |
| ACT Ontology v4 | |
| Improved patient counting scripts ("totalnum") | |
Synthea SyntheticMass dataset in i2b2 format | |
Simplified database upgrade method | |
log4J upgrade (to address security concerns) | |
Code changes to address security vulnerabilities | |
Bugfixes |
Community-Contributed Features
Contribution | Contributor | |
| SAML Authentication | Kevin Bui (lead developer) | i2b2 now includes support for SAML-based enterprise authentication via an institutional Identity Provider. See more information below. |
Ability to specify user parameter defaults | Michael Horvath | This change is meant to |
allow user params to take precedence over hive params. Currently, it's the other way around. |
In the process of making this change, I re-organized basic authentication into its own package and removed some code duplication.
| ||
LDAP UPN Support | Michael Horvath | Active Directory enables other methods of binding which are more flexible besides just using the distinguished name. https://docs.microsoft.com/en-us/openspecs/windows_protocols/ms-adts/6a5891b8-928e-4b75-a4a5-0e3b77eaca52. This change is to enable binding the the User Principle Name form, which is very convenient when the distinguished names for users is not easily available (OU by department, etc.). |
| API to get all children of an ontology node | Kevin Bui | The metadata GetChildren API call, which returns information on the children of an ontology node, can now be configured to return multiple levels of children (e.g., children, children's children, etc.). This is done by specifying the numLevel parameters. By default, the function assumes numLevel = 1 and will return the direct descendants of the concept, which is one level of children. When the numLevel = -1 the function will return ALL descendants of the concept, otherwise the function will return up to and including the number of levels specified by numLevel (eg. numlevel=2 returns two levels of descendants, numLevel=4 returns four levels of descendants). |
| Totalnum Counter Performance Improvements | Darren Henderson University of Kentucky | Performance enhancements on SQL Server totalnum counting |
| : stop unnecessarily |
| recomputing temp tables. |
Backend Features
SAML Authentication
(LINKS DON'T WORK AND DOCUMENTATION IS INCOMPLETE.)
| ACT v4 Postgres bugfixes | Ambreen Zaver | Bugfixes in time interval calculation (for age and age-at-visit) in ACT v4 ontology for Postgres. |
Detailed Documentation on New Features
User Registration Tool
There is a new user registration tool that can be enabled in the webclient. It allows users to request an i2b2 account that can then be activated by an administrator. It supports manual entry of user information through the form shown below, or automatic population of user information through SAML.
Documentation on this new feature is here: 6.5a i2b2 Webclient User Registration
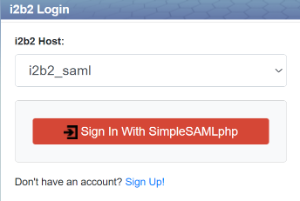
SAML Authentication
i2b2 now includes support for SAML-based enterprise authentication via an institutional Identity Provider. To configure this, you need to configure SimpleSAMLPHP (now included with i2b2) to talk to your institution's Identity Provider. To set up SAML:
We will use SimpleSAMLphp for IdP. Place the following files to the folder /etc/shibboleth/:
If you would like to use your own IdP, please visit Configuration - Service Provider 3 - Shibboleth Wiki for advance configurations.
Place the following files in the directory /etc/httpd/conf.d/:
1) Setting up Apache and simplesamlphp: https://simplesamlphp.org/docs/latest/simplesamlphp-install.html
2) Configure the service provider and add an identity provider: https://simplesamlphp.org/docs/latest/simplesamlphp-sp.html
(You will need to generate a cert in /var/www/simplesamlphp/metadata/saml20-idp-remote.php)
Detailed setup instructions are in Chapter 8 of the Installation Guide.
Improved Totalnum Scripts
Totalnum Scripts for Postgres and MSSQL (patient counting scripts) have been updated to improve the Totalnum counter's performance on both multiple ontology tables and very large(>1.5 million) ontology elements ontologies such as ACT medications, . Debug messages have also been added for troubleshoot purposestroubleshooting and profiling purposes. Support for multiple fact tables has been added and bugfixes have been made.
Totalnum Scripts Setup
- If upgrading, create the totalnum and totalnum_report tables. In Release_1-7/Upgrade/Metadata, run the ant upgrade script.
ant -f data_build.xml upgrade_tables_release_1-7-12a - In the Release_1-7/NewInstall/Metadata/ run the ant script to create the stored procedures.
...
-
ant -f data_build.xml create_metadata_procedures_release_1-
...
- 7
- Set privileges: If using multiple schemas, the stored procedure should be run from the metadata schema. Make sure the stored procedure can read the tables in the crcdata schema (observation_fact, visit_dimension, patient_dimension) and can both read an update ontology tables in the metadata schema (including table_access).
- If using multiple fact tables, the recommended approach is to create a fact table view as the union of all your fact tables. (This is essentially going back to a single fact table, but it is only used for totalnum counting. This is needed to correctly count patients that mention multiple fact tables within a hierarchy.)
e.g., create view observation_fact_view as select * from CONDITION_VIEW union all select * from drug_view
If running the counting script in SQL Server, add the wildcard flag, to ignore multifact references in the ontology: e.g. exec RunTotalnum 'observation_fact_view','dbo','@','Y'
This is automatically accounted for in the other database platforms. Note this approach does not work if you have conflicting concept_cds across fact tables. - Run the stored procedures on your database. This can be done in two ways
...
- :
...
- Run the ant command to execute the data_build.xml file with below specified target
POSTGRESQL : ant -f data_build.xml db_metadata_run_total_count_postgresql
ORACLE : ant -f data_build.xml db_metadata_run_total_count_oracle
SQL SERVER : ant -f data_build.xml db_metadata_run_total_count_sqlserver
- Run the ant command to execute the data_build.xml file with below specified target
...
- Execute the RunTotalNum stored procedure
...
- manually against your database
...
- from a
...
- SQL Client. This can take several hours
...
- for large databases or large ontologies. Examples are below.
Oracle: |
...
...
begin |
...
RUNTOTALNUM('observation_fact','i2b2demodata'); |
...
You can optionally include a table name if you only want to count one ontology table (this IS case sensitive): Note: If you get the error as: ERROR at line 1: ORA-01031: insufficient privilege, then run the command: |
...
grant create table to (DB USER) |
...
| SQL server: |
...
...
| exec RunTotalnum 'observation_fact','dbo','@' Parameters are: 1) the observation table name (for multi-fact-table setups), 2) the schema name, 3) a single table name to run on a single ontology table or '@' to run on all, and 4) and a wildcard flag that will ignore multifact references in the ontology if 'Y' | |
| PostgreSQL: | select RUNTOTALNUM('observation_fact','public') |
...
| Replace 'public' by the schema name for the fact table |
...
...
| If using a schema other than public for metadata, you might need to run "set search_path to 'i2b2metadata','public' " first as well |
...
When finished, verify it is complete by checking that c_totalnum columns in your ontology tables contain numbers (not nulls).
...
...
These total counts will be visible in the ontology browser in the web client.
Parent folders will get counts (of all patients with facts in the leaves) except for ontology folders derived from visit_dimension or patient_dimension. These cannot be rolled up because of the way these terms are defined in the ontology. They will have no count at all (not a zero).
| Info |
|---|
i2b2 users must have the DATA_AGG user permission to view the counts through the web client. |
Additional New Stored Procedures
Age In Years Updater
TODOWhen the CRC data is installed via ant, a new SQL script updates the age_in_years_num in the patient dimension based on the birth dates of the sample patients. As a reminder, this load process can be triggered with ant -f data_build.xml db_demodata_load_data in the CRC directory of NewInstall.
Concept Dimension Updater
TODO
Insert_Concept_FROMTableAccess is designed to populate concept_dimenison table using the ontologies listed in table_access table records.
The stored procedure loops through the table_access and inserts values from each metadata table (specified in the c_table_name column), when
c_dimtablename is set to 'concept_dimension'
Example usage: exec Insert_Concept_FROMTableAccess
I2b2-Synthea data Load
A new option is now available for loading Synthea data files into i2b2. Synthetic patient data generated by Synthea is hosted on SyntheticMass..can be loaded into i2b2. The Synthea SyntheticMass sample files have been converted to i2b2-ACT format. The zipped data files can be downloaded from , and scripts to load Synthea data from scratch are available here: https://github.com/i2b2/i2b2-synthea
Synthea Load Process
...
- Create db Schema with name labeled synthea
- Load Synthea data from the sample data files provided
- Download the zipped flat files and extract to them to a local directory
- Using SSMS, import the sample obs file into obvseration_fact table
- Import the sample obs-pat file into patient dimension table.
...
- Set up an i2b2 project with the ACT ontology.
- Either download the SyntheticMass 63k sample in i2b2 format from https://github.com/i2b2/i2b2-synthea/blob/main/syntheamass_63K_sample.zip, or follow the instructions below to load any Synthea dataset from scratch. This information can also be found on the Synthea-i2b2 Community Project page.
Loading Synthea data from scratch
- Download SyntheticMass Data, Version 2 (24 May, 2017)
- All data sets (1k, COVID 10k, COVID 100k) have been verified to work EXCEPT the 100k patients in the large SyntheticMass Version 2 download.
- The 100k patients in the large SyntheticMass Version 2 download needs an extra step to delete invalid records before import. In this case, download synthea_cleanup.pl to your disk, and then run "synthea_cleanup <directory-for-synthea-csv-files>" The fixed csv files will be in <directory-for-synthea-csv-files>/fixcsv.
- Set up an i2b2 project with the ACT ontology.
- Download the scripts from https://github.com/i2b2/i2b2-synthea
- Run
create_synthea_table_sqlserver<your dbServertype>.sqlin your project to create the Synthea tables. - Import the Synthea data you downloaded in step one into the Synthea tables in your project.
- Load the i2b2-to-SNOMED table in this repository into your project. https://www.nlm.nih.gov/healthit/snomedct/us_edition.html
- Click on the "Download SNOMED-CT to ICD-10-CM Mapping Resources" link to download. (You will need a UMLS account.)
- Unzip the file
- Import the TSV file into a table called SNOMED_to_ICD10 in your database.
- In Postgres and Oracle, follow the additional instructions in the comments at the top of
synthea_to_i2b2_<your dbServerType>.sqlto clean up the date formatting. - Run
synthea_to_i2b2_sqlserver<your dbServertype>.sqlto convert synthea Synthea data into i2b2 tables (this will truncate your existing fact and dimension tables!) - Replace references to
i2b2metadata.dboin the script. Use the database and schema where your ACT ontology tables are.
ACT Version-4 Ontology data load
Metadata scripts are now available to load the latest ACT Version-4 Ontology 4 ontology into your i2b2 db schema.
ACT4 dataload process:
...
| Note |
|---|
The CPT4 ontology table is not included with i2b2 due to AMA restrictions on redistribution of CPT code information. Contact the ACT team to get a copy if your institution is an AMA member. |
ACT4 data load process
- Download and extract the newinstall zip package from "Download Binary Distribution" in the top section of https://www.i2b2.org/software
...
- Edit the edu.harvard.i2b2.data\Release_1-7\NewInstall\Metadata\
...
- db.properties
...
- file to update the project properties to 'ACT' ; db.project=ACT
- From the edu.harvard.i2b2.data\Release_1-7\NewInstall\Metadata folder, run the ant command: ant -f data_build.xml db_metadata_load_data
- This will execute the SQL scripts
...
- from the edu.harvard.i2b2.data\Release_1-7\NewInstall\Metadata\act
...
- \scripts\<db type> folder and create and load ACT4 Ontology metadata tables
- You can now verify the new Ontology by logging into the webclient.
The following command will load the corresponding concept_dimension data of the Onbtology tables that will enable you to run queries in the webclient
From a sql Client>select 'insert into concept_dimension select c_dimcode AS concept_path, c_basecode AS concept_cd, c_name AS name_char, null AS concept_blob, update_date AS update_date, download_date as download_date, import_date as import_date, sourcesystem_cd as sourcesystem_cd, 1 as upload_id from '
+c_table_name+' where m_applied_path=''@'' and c_tablename=''CONCEPT_DIMENSION'' and c_columnname=''concept_path'' and c_visualattributes not like ''%I%'' and (c_columndatatype=''T'' or c_columndatatype=''N'') and c_synonym_cd = ''N'' and (m_exclusion_cd is null or m_exclusion_cd='''') and c_basecode is not null and c_basecode!='''''
from <your_dbschema>.dbo.TABLE_ACCESS where c_visualattributes like '%A%'
Improved db Upgrade Process
Currently i2b2 db upgrade is a multi-step process of running upgrade scripts and stored procedures. This release provides a set of upgrade scripts which will perform the complete db upgrade.
based on your current build version.
For example: Following Ant command will upgrade your db instance from 1.7.09c to latest version.
>ant -f data_build.xml upgrade_table_release_1-7-09c upgrade_table_release_1-7-10 upgrade_table_release_1-7-11 upgrade_table_release_1-7-12
Steps to Perform db upgrade:
- Backup your existing data folder
- Copy all the folders from the extracted download data folder into your existing data Upgrade folder
Example: Downloads\2b2core-upgrade-1712a\i2b2\data to C:\opt\edu.harvard.i2b2.data\Release_1-7\Upgrade\. This will replace
existing Crcdata, Hivedata, Metadata, PMdata folders.
Alternative to above step, navigate to the edu.harvard.i2b2.data\Release_1-7\Upgrade\ directory of your extracted folder - Copy the db.properties files from your back up into the respective locations(namely Crcdata, Hivedata, Metadata, PMdata )
- Open the command prompt and navigate to cell folders and run the following upgrade ant commands on your i2b2 database instance, where {db} can be Oracle, sqlserver or postgresql.
Alternative to above Step, you can run individual SQL scripts on your db instance in place of ant commands.
...
In data folder\Release_1-7\Upgrade\ run the ant commands under each individual cell subfolder as below.
...
1.7.09c
...
In the Crcdata folder run the following ant command: ant -f data_build.xml upgrade_table_release_1-7-09c upgrade_table_release_1-7-10 upgrade_table_release_1-7-11 upgrade_table_release_1-7-12
In the Hivedata folder run the following ant command: ant -f data_build.xml upgrade_hive_tables_release_1-7-09c upgrade_hive_tables_release_1-7-10 upgrade_hive_tables_release_1-7-11 upgrade_hive_tables_release_1-7-12
In the Metadata folder run the following ant command: ant -f data_build.xml upgrade_tables_release_1-7-09c upgrade_tables_release_1-7-10 upgrade_tables_release_1-7-11 upgrade_tables_release_1-7-12
In the PMdata folder run the following ant command: ant -f data_build.xml upgrade_pm_tables_release_1-7-09c upgrade_pm_tables_release_1-7-10 upgrade_pm_tables_release_1-7-11 upgrade_pm_tables_release_1-7-12
...
1.7.10
...
In the Crcdata folder run the following ant command: ant -f data_build.xml upgrade_table_release_1-7-10 upgrade_table_release_1-7-11 upgrade_table_release_1-7-12
In the Hivedata folder run the following ant command: ant -f data_build.xml upgrade_hive_tables_release_1-7-10 upgrade_hive_tables_release_1-7-11 upgrade_hive_tables_release_1-7-12
In the Metadata folder run the following ant command: ant -f data_build.xml upgrade_tables_release_1-7-10 upgrade_tables_release_1-7-11 upgrade_tables_release_1-7-12
In the PMdata folder run the following ant command: ant -f data_build.xml upgrade_pm_tables_release_1-7-10 upgrade_pm_tables_release_1-7-11 upgrade_pm_tables_release_1-7-12
1.7.11
...
In the Crcdata folder run the following ant command: ant -f data_build.xml upgrade_table_release_1-7-11 upgrade_table_release_1-7-12
In the Hivedata folder run the following ant command: ant -f data_build.xml upgrade_hive_tables_release_1-7-11 upgrade_hive_tables_release_1-7-12
In the Metadata folder run the following ant command: ant -f data_build.xml upgrade_tables_release_1-7-11 upgrade_tables_release_1-7-12
In the PMdata folder run the following ant command: ant -f data_build.xml upgrade_pm_tables_release_1-7-11 upgrade_pm_tables_release_1-7-12
...
In the Crcdata folder run the following ant command: ant -f data_build.xml upgrade_table_release_1-7-12
In the Hivedata folder run the following ant command: ant -f data_build.xml upgrade_hive_tables_release_1-7-12
In the Metadata folder run the following ant command: ant -f data_build.xml upgrade_tables_release_1-7-12
In the PMdata folder run the following ant command: ant -f data_build.xml upgrade_pm_tables_release_1-7-12
Security Enhancements
- i2b2 has been made more secure by addressing parameterization and other potential vulnerabilities found in an internal a Veracode scan.
- Log4J has been upgraded to the latest version. The following jars are updated in lib/axis2.war/WEB-INF/lib the folder:
- log4j-api-2.17.1.jar
- log4j-core-2.17.1.jar
- log4j-jcl-2.17.1.jar
Improved db Upgrade Process
Previously, i2b2 db upgrade was a multi-step process of running upgrade SQL scripts and stored procedures individually on the db instance. This release simplifies the process of running the table upgrade SQL scripts and stored procedures from data_build.xml files. Details are on the i2b2 Upgrade Page here.
Changelog
Database Drivers
The JDBC drivers were updated to the following versions.
Driver | ojdbc8.jar | postgresql-42.2. |
|---|
14.jar | mssql-jdbc- |
9. |
2.0.jre8.jar | |
New Version | Oracle |
|---|
21.5 | PostgreSQL 42.3.2 |
MS Sql Server |
9. |
2 |
Supported Db Server versions
Server Type | SQL Server | Oracle | Postgres |
|---|---|---|---|
Supported Version/s | 2012+ (tested with up to 2019) | 12g+ and 21c | 9 to 14 |
Supported software versions
Application Type | Java | Wildfly | Apache HTD | Apache Ant | Apache Axis2 | PHP |
|---|---|---|---|---|---|---|
Supported Version/s | 8 or 11 | 17.0. |
1Final | 2. |
0 (RHEL 6) and 2.2 ( RHEL 7) | 1.9.6 | 1.7.1 | 7.2.27 or higher |
i2b2 Database Changes
...
- DATA-7 QT_PATIENT_SET_ENC_COLLECTION should be a bigint
- DATA-5 Synthea i2b2 data
- DATA-8 Synthea data load test from Github files
- DATA-6 improve i2b2 db upgrade process
- DATA-12 Stored procedure to update concept dimension
- DATA-11 Age in years updater during demodata install
- DATA-9 ACT v4 ontology
- CORE-389 Totalnum performance improvements
- CORE-394 Obfuscated totalnum reporting tables
- CORE-398 Multifact support for totalnums
- CORE-400 Show totalnums in top level folders
Supported Operating Systems
CentOS versions 6 (deprecated) or 7 (highly recommended)
Windows 7-2019
Unofficially, MacOS and other flavors of Linux are likely to work.
i2b2 Server and Client Changes
New Features and Improvements
| Core- |
|---|
- WEBCLIENT-334 Provide tabs for major plugins and temporal query
- WEBCLIENT-344 Cleanup Analysis Tools list of Plugins to only Supported Items
- WEBCLIENT-325 Wayne's improvements to hierarchical find-by-name
- WEBCLIENT-320 Provide REFRESH ALL context menu in FindTerms panel
- WEBCLIENT-307 Drag and drop in term info panel
- WEBCLIENT-306 Beth Israel contribution: commas in counts, query option infrastructure
- WEBCLIENT-343 Fix Veracode identified Security flaws in i2b2Webclient Code
- WEBCLIENT-353 SAML (client side)
| server | webclient |
|---|---|
CORE-399 Oracle index hints must use the table alias CORE-382 Username / password errors should not specify which had the problem CORE-402 Fix Veracode identified Security flaws in i2b2 Server-Side Code CORE-404 Adding support for JDK 11. Now including the gensrc due to jaxb has been removed. |
WEBCLIENT-334 Provide tabs for major plugins and temporal query WEBCLIENT-344 Cleanup Analysis Tools list of Plugins to only Supported Items WEBCLIENT-325 Wayne's improvements to hierarchical find-by-name WEBCLIENT-353 SAML and user registration tool (client side) |
i2b2 Database Changes
New Features and Improvements
DATA-7 QT_PATIENT_SET_ENC_COLLECTION should be a bigint DATA-14 Synthea i2b2 data DATA-6 improve i2b2 db upgrade process DATA-12 Stored procedure to update concept dimension DATA-11 Age in years updater during demodata install DATA-9 ACT v4 ontology DATA-13 Postgres time interval corrections in ACT v4 demographics ontology CORE-389 Totalnum performance improvements CORE-394 Obfuscated totalnum reporting table CORE-398 Multifact support for totalnums CORE-400 Show totalnums in top level folders |
Bug Fixes
| Webclient |
|---|
| Core- |
|---|
| server |
|---|
WEBCLIENT-351 Obfuscated User Not Showing Graph WEBCLIENT-342 switch response status check from "OK" to 200 to handle lab value pop up in http/2 protocols WEBCLIENT-335 temporal query in webclient with no anchoring events not running WEBCLIENT-350 Unable to drag items in |
workplace |
294 Webclient Reports "QUERY CANCELLED" While Query Is Still |
Running |
354 Removed broken context menu in Find Previous Queries | CORE-418 Local timestamps in sessions on Oracle - appserver /db server in diff time |
zones CORE-282 Error returned when obfuscated user is locked |
out CORE-281 Query continues to run after user receives lockout |
message |
399 use alias for index |
hint |
Notes for Developers
For Java 11 install, if you change the xsd , then modify the gensource(REST API message definitions), then you will need to regenerate gensrc via JAXB in Java 8. In the i2b2-core cell directory for which you're regenerating the XSD-Java, run the ant target "jaxb_gen" on Java 8 and then build as usual using Java 11.
...







.svg.med.png)
