Last Updated: 03/19/2013
Medical history: diagnosis modifier usability [01-31-2013]
From: Dan Connolly [dconnolly.kumc] Sent: Thursday, January 31, 2013 10:00 AM To: members Cc: Matthew Hoag; Tamara McMahon Subject: medical history: diagnosis modifier usability? alternatives?
We recently integrated medical history data into HERON, our i2b2 installation. In our hospital EMR, medical histories are represented with ICD9 codes, so we put them in i2b2 with concept_cd=ICD9:xyz, with a "Medical History diagnosis" modifier. I wonder about the usability of this approach and if anyone has explore other alternatives. To illustrate: if I query for, say, hypertensive disease (ICD9:401-405.99) in the last 3 years or so, for any clinical diagnosis modifier, I get about 63K patients. But if I do the same query dragging in each of the clinical diagnosis modifiers (encounter diagnosis, problem list, ...; see attached screenshot) except medical history, the count goes down to 46K. Our hypertensive disease concept actually means "diagnosed with hypertensive disease or reported a history of hypertensive disease" now. And if a researcher wants just "diagnosed with hypertensive disease," they have to drag in 5 modifiers under hypertensive disease, rather than just dragging in hypertensive disease. I think it's fairly common for researchers to query for dozens of ICD9 codes. Doing this modifier exercise dozens of times seems like an awful lot to ask. I suppose another alternative is to use one Problem status=Active modifier (as opposed to Deleted or Resolved); I get 24K in that case. We also have a primary billing modifier; I get 6K in that case. Looking for both (i.e. either active or primary billing) gives 27K, so there's some overlap. If anyone has other experience to share related to medical history data, I'd appreciate hearing about it.
Dan ConnollyBiomedical Informatics DivisionKU Med Center
Shibboleth wrapping of i2b2 1.6 client [01-29-2013]
From: Dat Q. Phan [Dat.Q.Phan.uth.tmc] Sent: Tuesday, January 29, 2013 10:30 AM To: Nick Anderson; i2b2 AUG Members Cc: Elmer V Bernstam Subject: RE: Shibboleth wrapping of i2b2 1.6 client?
Nick,
We currently use Shibboleth authentication with i2b2 v1.6 for our Integrated Dental Data Repository at UTHealth. Let me know what I can help you with.
Regards,
Johnny Phan
Programmer Analyst
UTHealth
School of Biomedical Informatics
Dat.Q.Phan.uth.tmc
From: Russ Waitman [rwaitman.kumc] Sent: Sunday, January 27, 2013 12:28 PM To: Nick Anderson; i2b2 AUG Members Cc: Dan Connolly Subject: RE: Shibboleth wrapping of i2b2 1.6 client?
Hi Nick, Dan Connolly did similar work to auto-authenticate based on CAS/LDAP integration but not Shib. We may get there. The spots in the code though I bet are very very similar and I remember 2 years ago Dan Connolly meeting with Philip and Mike Mendis at the i2b2 conference on the topic. The code's on our wiki probably where we made PM modshttps://informatics.kumc.edu/work/browser/i2b2-pm But Dan will know for sure,Russ
From: Nick Anderson [nicka.u.washington] Sent: Thursday, January 24, 2013 2:20 PM To: i2b2 AUG Members Subject: Shibboleth wrapping of i2b2 1.6 client?
Has anybody successfully managed to wrap the 1.6 client to allow for direct Shibboleth authentication? We had it for 1.3 via Phil Reeders helpful work, and would rather gain from external experience than develop it again de novo.
thanks! - Nick
Nick Anderson, Ph.D.Assistant Professor, Department of Biomedical Informatics and Medical EducationAdjunct Assistant Professor, Bioethics and HumanitiesAssociate Director, Biomedical Informatics Core, Institute of Translational Health Sciences
eScience InstituteUniversity of Washington, Seattle, Washington nicka.uwfaculty.washington.edu/nicka(206) 685 0249
Non-null C_TOOLTIP Path Required for Web Client v1.6.08 [01-25-2013]
From: Jack London [Jack.London.jefferson] Sent: Friday, January 25, 2013 9:56 AMTo: Murphy, Shawn N.Cc: i2b2 AUG MembersSubject: Re: non-null c_tooltip path required for web client v1.06.08?
Shawn,
I just set the c_tooltip column equal to the c_fullname where the c_tooltip column was null as a quick work-around. This is on a new development server VM we are setting up (hence the installation of the latest i2b2 version). Currently the i2b2 database on this development VM is "in flux" (we are loading new data, etc.). I will experiment with a "blank space" rather than a "null" next week and let you know what happens.
Jack
On Jan 25, 2013, at 7:02 AM, Murphy, Shawn N. wrote:
We'll take care of this in the next release Jack. A similar behavior on the Workbench will also be fixed. In the mean time, did you find a blank or space in the tooltip field fixes the problem?
Thanks,
Shawn.
From: Jack London [Jack.London.jefferson]
Sent: Thursday, January 24, 2013 2:50 PM
To: i2b2 AUG Members
Subject: non-null c_tooltip path required for web client v1.06.08?
I had always considered the c_tooltip column in the i2b2 metadata tables as optional.
On installing web client v1.06.08 I noticed that for some metadata selections, when I hit the "run query" button, there would sometimes not be anything in the "query name" field, or the previous query name would appear. In either event, nothing would happen – the client never made a query request to the server (no MASTER_QUERY table entries created).
On further exploration, I determined that this ONLY occurred if a data element was selected for which the metadata table C_TOOLTIP column was null. Furthermore, the problem was fixed after I entered text for this column.
Anyone else experience this?
Jack
V1.6.04 Web Client – CRC Queues handoff – Query status Panel – keeps running [01-24-2013]
From: Luke Rasmussen [luke.rasmussen.northwestern] Sent: Thursday, January 24, 2013 11:56 AM To: Peter Beninato; i2b2-install-help.googlegroups; 'i2b2 AUG Members' Subject: Re: i2b2 - v1.6.04 - WebClient - CRC Queues handoff - Query status Panel - keeps running
Peter – I apologize for my vague attempt at helping, but I do recall something like that happening before. Try Chrome's dev tools or Firebug to watch the communications to the web server. I believe the issue we had was that within the proxy page the call to the web service was timing out. In the browser I would see a 500 (error) response back from the proxy page request, and was able to track it down from there. Please note that we ported the original PHP proxy page to an ASP.NET MVC version, so it's possible the issues we had are unrelated.
Hopefully looking at the client will give you a hint. I'll see if I can reproduce the issue again on our side and let you know if I come across any other ideas.
Luke Rasmussen
Systems Analyst/Programmer
Northwestern University
Division of Health and Biomedical Informatics
(312) 503-2823
From: Peter Beninato <beninato.ohsu> Date: Thursday, January 24, 2013 10:39 AM To: "i2b2-install-help.googlegroups" <i2b2-install-help.googlegroups>, i2b2 AUG Members Subject: i2b2 - v1.6.04 - WebClient - CRC Queues handoff - Query status Panel - keeps running
Hi,
Trying to diagnose and solve this problem. When a query runs long enough to transition from the small to medium queue, the Query Status window continues to show that the query is running.
Eventually, the query runs to completion, but it doesnot seem that this status is communicated back to webclient Query Status window. It seems to be necessary to logout, and log back in.
Below is a stack trace, from around the time of the queue handoff.
Have others experienced this application behavior? How has it been resolved?
Is there some additional JBOSS configuration required?
Thanks.
2013-01-23 16:18:27,706 INFO [edu.harvard.i2b2.crc.dao.OracleDAOFactory] Using datasource java:QueryToolcohortsDS
2013-01-23 16:20:51,356 INFO [edu.harvard.i2b2.crc.ejb.QueryManagerBeanUtil] STATUS IS RUNNING RUNNING
2013-01-23 16:20:51,724 INFO [edu.harvard.i2b2.crc.dao.DataSourceLookupHelper] Located DataSource for hiveId=[octri] projectId=[/cohorts/]
2013-01-23 16:20:51,724 INFO [edu.harvard.i2b2.crc.dao.OracleDAOFactory] Using datasource java:QueryToolcohortsDS
2013-01-23 16:22:59,076 WARN [org.jboss.resource.connectionmanager.TxConnectionManager] Connection error occured: org.jboss.resource.connectionmanager.TxConnectionManager$TxConnectionEventListener@dd8cb4[state=NORMAL mc=org.jboss.resource.adapter.jdbc.local.LocalManagedConnection@1210e0e handles=1 lastUse=1358986707705 permit=true trackByTx=false mcp=org.jboss.resource.connectionmanager.JBossManagedConnectionPool$OnePool@91f904 context=org.jboss.resource.connectionmanager.InternalManagedConnectionPool@173b47a xaResource=org.jboss.resource.connectionmanager.TxConnectionManager$LocalXAResource@1409575 txSync=null]
java.sql.SQLException: ORA-01013: user requested cancel of current operation
at oracle.jdbc.driver.DatabaseError.throwSqlException(DatabaseError.java:112)
at oracle.jdbc.driver.T4CTTIoer.processError(T4CTTIoer.java:331)
at oracle.jdbc.driver.T4CTTIoer.processError(T4CTTIoer.java:288)
at oracle.jdbc.driver.T4C8Oall.receive(T4C8Oall.java:743)
at oracle.jdbc.driver.T4CStatement.doOall8(T4CStatement.java:207)
at oracle.jdbc.driver.T4CStatement.executeForRows(T4CStatement.java:946)
at oracle.jdbc.driver.OracleStatement.doExecuteWithTimeout(OracleStatement.java:1168)
at oracle.jdbc.driver.OracleStatement.executeUpdateInternal(OracleStatement.java:1614)
at oracle.jdbc.driver.OracleStatement.executeUpdate(OracleStatement.java:1579)
at org.jboss.resource.adapter.jdbc.WrappedStatement.executeUpdate(WrappedStatement.java:186)
at edu.harvard.i2b2.crc.dao.setfinder.QueryExecutorHelperDao.executeQuery(Unknown Source)
at edu.harvard.i2b2.crc.dao.setfinder.QueryExecutorDao.executeSQL(Unknown Source)
at edu.harvard.i2b2.crc.ejb.ExecRunnable.processQueryRequest(Unknown Source)
at edu.harvard.i2b2.crc.ejb.ExecRunnable.execute(Unknown Source)
at edu.harvard.i2b2.crc.ejb.QueryExecutorMDB.onMessage(Unknown Source)
at edu.harvard.i2b2.crc.ejb.QuerySmallExecutorMDB.onMessage(Unknown Source)
at sun.reflect.GeneratedMethodAccessor291.invoke(Unknown Source)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:25)
at java.lang.reflect.Method.invoke(Method.java:597)
at org.jboss.invocation.Invocation.performCall(Invocation.java:359)
at org.jboss.ejb.MessageDrivenContainer$ContainerInterceptor.invoke(MessageDrivenContainer.java:495)
at org.jboss.resource.connectionmanager.CachedConnectionInterceptor.invoke(CachedConnectionInterceptor.java:158)
at org.jboss.ejb.plugins.CallValidationInterceptor.invoke(CallValidationInterceptor.java:63)
at org.jboss.ejb.plugins.AbstractTxInterceptor.invokeNext(AbstractTxInterceptor.java:121)
at org.jboss.ejb.plugins.AbstractTxInterceptorBMT.invokeNext(AbstractTxInterceptorBMT.java:173)
at org.jboss.ejb.plugins.MessageDrivenTxInterceptorBMT.invoke(MessageDrivenTxInterceptorBMT.java:47)
at org.jboss.ejb.plugins.MessageDrivenInstanceInterceptor.invoke(MessageDrivenInstanceInterceptor.java:116)
at org.jboss.ejb.plugins.RunAsSecurityInterceptor.invoke(RunAsSecurityInterceptor.java:109)
at org.jboss.ejb.plugins.LogInterceptor.invoke(LogInterceptor.java:205)
at org.jboss.ejb.plugins.ProxyFactoryFinderInterceptor.invoke(ProxyFactoryFinderInterceptor.java:138)
at org.jboss.ejb.MessageDrivenContainer.internalInvoke(MessageDrivenContainer.java:402)
at org.jboss.ejb.Container.invoke(Container.java:960)
at org.jboss.ejb.plugins.jms.JMSContainerInvoker.invoke(JMSContainerInvoker.java:987)
at org.jboss.ejb.plugins.jms.JMSContainerInvoker$MessageListenerImpl.onMessage(JMSContainerInvoker.java:1287)
at org.jboss.jms.asf.StdServerSession.onMessage(StdServerSession.java:266)
at org.jboss.mq.SpyMessageConsumer.sessionConsumerProcessMessage(SpyMessageConsumer.java:891)
at org.jboss.mq.SpyMessageConsumer.addMessage(SpyMessageConsumer.java:170)
at org.jboss.mq.SpySession.run(SpySession.java:323)
at org.jboss.jms.asf.StdServerSession.run(StdServerSession.java:194)
at EDU.oswego.cs.dl.util.concurrent.PooledExecutor$Worker.run(PooledExecutor.java:761)
at java.lang.Thread.run(Thread.java:619)
2013-01-23 16:22:59,095 ERROR [edu.harvard.i2b2.crc.ejb.ExecRunnable] ORA-01013: user requested cancel of current operation
edu.harvard.i2b2.crc.dao.setfinder.CRCTimeOutException: ORA-01013: user requested cancel of current operation
at edu.harvard.i2b2.crc.dao.setfinder.QueryExecutorHelperDao.executeQuery(Unknown Source)
at edu.harvard.i2b2.crc.dao.setfinder.QueryExecutorDao.executeSQL(Unknown Source)
at edu.harvard.i2b2.crc.ejb.ExecRunnable.processQueryRequest(Unknown Source)
at edu.harvard.i2b2.crc.ejb.ExecRunnable.execute(Unknown Source)
at edu.harvard.i2b2.crc.ejb.QueryExecutorMDB.onMessage(Unknown Source)
at edu.harvard.i2b2.crc.ejb.QuerySmallExecutorMDB.onMessage(Unknown Source)
at sun.reflect.GeneratedMethodAccessor291.invoke(Unknown Source)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:25)
at java.lang.reflect.Method.invoke(Method.java:597)
at org.jboss.invocation.Invocation.performCall(Invocation.java:359)
at org.jboss.ejb.MessageDrivenContainer$ContainerInterceptor.invoke(MessageDrivenContainer.java:495)
at org.jboss.resource.connectionmanager.CachedConnectionInterceptor.invoke(CachedConnectionInterceptor.java:158)
at org.jboss.ejb.plugins.CallValidationInterceptor.invoke(CallValidationInterceptor.java:63)
at org.jboss.ejb.plugins.AbstractTxInterceptor.invokeNext(AbstractTxInterceptor.java:121)
at org.jboss.ejb.plugins.AbstractTxInterceptorBMT.invokeNext(AbstractTxInterceptorBMT.java:173)
at org.jboss.ejb.plugins.MessageDrivenTxInterceptorBMT.invoke(MessageDrivenTxInterceptorBMT.java:47)
at org.jboss.ejb.plugins.MessageDrivenInstanceInterceptor.invoke(MessageDrivenInstanceInterceptor.java:116)
at org.jboss.ejb.plugins.RunAsSecurityInterceptor.invoke(RunAsSecurityInterceptor.java:109)
at org.jboss.ejb.plugins.LogInterceptor.invoke(LogInterceptor.java:205)
at org.jboss.ejb.plugins.ProxyFactoryFinderInterceptor.invoke(ProxyFactoryFinderInterceptor.java:138)
at org.jboss.ejb.MessageDrivenContainer.internalInvoke(MessageDrivenContainer.java:402)
at org.jboss.ejb.Container.invoke(Container.java:960)
at org.jboss.ejb.plugins.jms.JMSContainerInvoker.invoke(JMSContainerInvoker.java:987)
at org.jboss.ejb.plugins.jms.JMSContainerInvoker$MessageListenerImpl.onMessage(JMSContainerInvoker.java:1287)
at org.jboss.jms.asf.StdServerSession.onMessage(StdServerSession.java:266)
at org.jboss.mq.SpyMessageConsumer.sessionConsumerProcessMessage(SpyMessageConsumer.java:891)
at org.jboss.mq.SpyMessageConsumer.addMessage(SpyMessageConsumer.java:170)
at org.jboss.mq.SpySession.run(SpySession.java:323)
at org.jboss.jms.asf.StdServerSession.run(StdServerSession.java:194)
at EDU.oswego.cs.dl.util.concurrent.PooledExecutor$Worker.run(PooledExecutor.java:761)
at java.lang.Thread.run(Thread.java:619)
Caused by: java.sql.SQLException: ORA-01013: user requested cancel of current operation
at oracle.jdbc.driver.DatabaseError.throwSqlException(DatabaseError.java:112)
at oracle.jdbc.driver.T4CTTIoer.processError(T4CTTIoer.java:331)
at oracle.jdbc.driver.T4CTTIoer.processError(T4CTTIoer.java:288)
at oracle.jdbc.driver.T4C8Oall.receive(T4C8Oall.java:743)
at oracle.jdbc.driver.T4CStatement.doOall8(T4CStatement.java:207)
at oracle.jdbc.driver.T4CStatement.executeForRows(T4CStatement.java:946)
at oracle.jdbc.driver.OracleStatement.doExecuteWithTimeout(OracleStatement.java:1168)
at oracle.jdbc.driver.OracleStatement.executeUpdateInternal(OracleStatement.java:1614)
at oracle.jdbc.driver.OracleStatement.executeUpdate(OracleStatement.java:1579)
at org.jboss.resource.adapter.jdbc.WrappedStatement.executeUpdate(WrappedStatement.java:186)
... 31 more
2013-01-23 16:22:59,658 INFO [edu.harvard.i2b2.crc.ejb.QueryMediumExecutorMDB] Executing from Medium queue for query instance [ 5149 ]
2013-01-23 16:22:59,675 INFO [edu.harvard.i2b2.crc.dao.DataSourceLookupHelper] Located DataSource for hiveId=[octri] projectId=[/cohorts/]
2013-01-23 16:22:59,675 INFO [edu.harvard.i2b2.crc.dao.OracleDAOFactory] Using datasource java:QueryToolcohortsDS
2013-01-23 16:23:00,399 INFO [edu.harvard.i2b2.crc.dao.DataSourceLookupHelper] Located DataSource for hiveId=[octri] projectId=[/cohorts/] and ownerId =[@]
2013-01-23 16:23:00,399 INFO [edu.harvard.i2b2.crc.dao.OracleDAOFactory] Using datasource java:QueryToolcohortsDS
Peter Beninato - OCTRI DW Developer
Oregon Health & Science University
503-494-9985
beninato.ohsu
Help with Workbench error message [01-15-2013]
From: Lee, Eunjung S [sallylee.u.washington] Sent: Tuesday, January 15, 2013 11:39 AM To: Phillips, Lori C.; Nick Anderson Subject: RE: Help with Workbench error message
Hi Lori,
It is strange that such error would pop up due to having no tooltip.
I just populated the tooltip for all missing ones and tried one mini-test on workbench (not sure if my copy really works well) and I got no error so far.
Nick,
Do you think you could let Lori know if workbench works now?
Thanks!
Sally
From: Phillips, Lori C. [LCPHILLIPS.PARTNERS] Sent: Tuesday, January 15, 2013 7:17 AM To: Nick Anderson Cc: E. S. Lee Subject: RE: Help with Workbench error message
It looks like its complaining because there is no tooltip.
From: Nick Anderson [nicka.u.washington] Sent: Monday, January 14, 2013 4:26 PM To: Phillips, Lori C. Cc: E. S. Lee Subject: Re: Help with Workbench error message
Yep - see below examples from both problem trees..
On Jan 14, 2013, at 1:13 PM, "Phillips, Lori C." <LCPHILLIPS.partners> wrote:
You can drag the concept (as text) to any window that accepts text.
<?xml version="1.0" encoding="UTF-8" standalone="yes"?>
<ns2:plugin_drag_drop xmlns:ns2="http://www.i2b2.org/xsd/hive/plugin/" xmlns:ns4="http://www.i2b2.org/xsd/hive/msg/1.1/" xmlns:ns3="http://www.i2b2.org/xsd/cell/crc/psm/1.1/" xmlns:ns5="http://www.i2b2.org/xsd/hive/msg/result/1.1/" xmlns:ns6="http://www.i2b2.org/xsd/cell/ont/1.1/" xmlns:ns7="http://www.i2b2.org/xsd/cell/crc/psm/querydefinition/1.1/">
<ns6:concepts>
<concept>
<level>4</level>
<key>\\i2b2\i2b2\Vitals\BloodPressure\Diastolic\60-79\</key>
<name> 60-79</name>
<synonym_cd>N</synonym_cd>
<visualattributes>FA</visualattributes>
<totalnum xsi:nil="true" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"/>
<facttablecolumn>concept_cd</facttablecolumn>
<tablename>concept_dimension</tablename>
<columnname>concept_path</columnname>
<columndatatype>T</columndatatype>
<operator>LIKE</operator>
<dimcode>\i2b2\Vitals\BloodPressure\Diastolic\60-79\</dimcode>
</concept>
</ns6:concepts>
</ns2:plugin_drag_drop>
<?xml version="1.0" encoding="UTF-8" standalone="yes"?>
<ns2:plugin_drag_drop xmlns:ns2="http://www.i2b2.org/xsd/hive/plugin/" xmlns:ns4="http://www.i2b2.org/xsd/hive/msg/1.1/" xmlns:ns3="http://www.i2b2.org/xsd/cell/crc/psm/1.1/" xmlns:ns5="http://www.i2b2.org/xsd/hive/msg/result/1.1/" xmlns:ns6="http://www.i2b2.org/xsd/cell/ont/1.1/" xmlns:ns7="http://www.i2b2.org/xsd/cell/crc/psm/querydefinition/1.1/">
<ns6:concepts>
<concept>
<level>5</level>
<key>\\i2b2\i2b2\Lab\Chemistry\Blood Electrolytes\BUN\LT8\</key>
<name> <8 mg/dL</name>
<synonym_cd>N</synonym_cd>
<visualattributes>LA</visualattributes>
<totalnum>7862</totalnum>
<basecode>LAB|BUN:<8</basecode>
<facttablecolumn>concept_cd</facttablecolumn>
<tablename>concept_dimension</tablename>
<columnname>concept_path</columnname>
<columndatatype>T</columndatatype>
<operator>LIKE</operator>
<dimcode>\i2b2\Lab\Chemistry\Blood Electrolytes\BUN\LT8\</dimcode>
</concept>
</ns6:concepts>
</ns2:plugin_drag_drop>
From: Phillips, Lori C. [LCPHILLIPS.PARTNERS] Sent: Monday, January 14, 2013 1:07 PM To: Lee, Eunjung S Subject: RE: Help with Workbench error message
Drag and drop the concept term directly to email ...
I will need to look at the info contained in the drag and drop.
Lori
From: Lee, Eunjung S [sallylee.u.washington] Sent: Monday, January 14, 2013 3:55 PM To: Phillips, Lori C. Cc: i2b2 AUG Members Subject: RE: Help with Workbench error message
Hi Lori,
I have already encountered that issue and fixed accordingly.
In c_fullname, there is no < in the pathname any longer (only shows on c_name, which should not be a problem).
Also, having < in the c_fullname caused error in the webclient as well.
I forgot to mention the workbench version is 1.6.05.0001 and the user who is using it is using OSX (however, same error was encountered also on windows workbench).
From: Phillips, Lori C. [LCPHILLIPS.PARTNERS] Sent: Monday, January 14, 2013 12:51 PM To: Lee, Eunjung S Cc: i2b2 AUG Members Subject: RE: Help with Workbench error message
Sally,
I think the problem is that the name of the term starts with "<" which the workbench may interpret as the beginning of an xml <tag>.
Is the problem isolated to terms of this type? Try renaming the term and its c_fullpath so that it reads "Less than"
Lori
From: Lee, Eunjung S [sallylee.u.washington] Sent: Monday, January 14, 2013 3:41 PM To: Phillips, Lori C. Cc: i2b2 AUG Members Subject: Re: Help with Workbench error message
Attached is the screen shot of an error given to me by a user who's working with the i2b2 workbench.
The error message is given off when querying 2 different Categories of our ontology (Lab and Vitals), both of which were built by us.
All of the queries that are giving error on workbench works fine on the webclient so we don't think it is an issue with the ontology itself.
We are thinking it is some type of permission setting?
Thanks for your help!
Yours,
Sally
From: Phillips, Lori C. [LCPHILLIPS.PARTNERS] Sent: Monday, January 14, 2013 9:04 AM To: Lee, Eunjung S Cc: i2b2 AUG Members Subject: RE: Help with Workbench error message
I will need more info about the item you are dragging.
Can you enclose a screenshot showing the term?
Will need to know i2b2 version info as well.
Thanks
Lori Phillips
From: Lee, Eunjung S [sallylee.u.washington] Sent: Monday, January 14, 2013 11:46 AM To: Phillips, Lori C.; Lee, Eunjung S Cc: i2b2 AUG Members Subject: RE: Help with Workbench error message
We are dragging leaf items and getting the error. Also, container item would not work on webclient either. But these are node items that work to return query on webclient but giving error on the workbench. Sally
"Phillips, Lori C." wrote:
Sally,
Are you trying to drag in a container? Containers are typically the highest level in the ontology tree.
They are not allowed to be dropped into the query tool. Open the container and select a sub category to query by.
Lori
From: Lee, Eunjung S [sallylee.u.washington] Sent: Monday, January 14, 2013 11:32 AM To: i2b2 AUG Members Subject: Help with Workbench error message
We are consistently getting an error message when using i2b2 workbench while trying to drag ontology items into the query tool area.
The message says: "Please note, You can not drop this item here" and it will not allow the items to be dragged to the query tool.
Since same type of query works on the webclient, we have no idea how to solve the problem.
The issue is, we do need to use the workbench tool to do some analysis for one of our projects so we need to be able to query using the workbench. We are hoping someone had faced the same issue before and could help us solve the problem.
Thanks!
Sally
E. Sally Lee, Ph.D
Biomedical Informatics Core Consult
Amalga Clinical Data Repository
Institute of Translational Health Sciences
University of Washington
sallylee.uw
Please help us continue to support your research by +citing our grant number +in publications we supported.
The ITHS is part of the national Clinical and Translational Science Awards (CTSA) consortium, and is funded under grant 2UL1TR000423-06 from the National Center for Research Resources and the National Center for Advancing Translational Sciences, part of the National Institutes of Health.
Mapping family history to a national standard [01-11-2013]
From: Tamara McMahon [tmcmahon.kumc] Sent: Friday, January 11, 2013 1:37 PM To: i2b2 AUG Members Subject: Mapping family history to a national standard
Hi,
We are in the process of pulling family history into our i2b2 instance. In our EMR (Epic) a pick list exists of medical/family history terms (around 370-400 terms). The concepts are listed but not in a standard ontology.
As a result, we are embarking on a project to map medical history to a national standard, preferably Snomed.
Have any Epic users in this group mapped these terms? If so, any desire to share advice or mappings? If not, any interest in collaborating?
All input is appreciated.
Tamara McMahon
Clinical Informatics Coordinator
Department of Biostatistics
University of Kansas Medical Center
913-945-7470
PDO Web Services and Adding Colmuns to the OBSERVATION_FACT Table [01-10-2013]
From: Murphy, Shawn N. Sent: Thursday, January 10, 2013 7:58 AM To: Dat Q. Phan; i2b2 AUG Members Subject: RE: PDO Web Services and Adding Columns to the Observation_Fact Table
Hi Johnny,
Perhaps best added as a modifier. There is no out-of-the-box configurability of the observation_fact columns like there are in the patient_dimension and the visit_dimension.
Thanks,
Shawn.
From: Dat Q. Phan [Dat.Q.Phan.uth.tmc] Sent: Wednesday, January 09, 2013 7:05 PM To: i2b2 AUG Members Subject: PDO Web Services and Adding Columns to the Observation_Fact Table
Hi,
I'm using the CRC Messaging to query i2b2 to return data from the Observation_Fact table.
In the documentation "CRC_Messaging.pdf", the "observation_set" includes the data:
<observation>
<event_id source="source3">event_id3</event_id>
<patient_id>patient_id9</patient_id>
<concept_cd name="name0">concept_cd3</concept_cd>
<observer_cd soruce="soruce0">observer_cd3</observer_cd>
<start_date>2006-05-04T18:13:51.0Z</start_date>
<modifier_cd name="name1">modifier_cd0</modifier_cd>
<instance_num>1</intance_num>
<valuetype_cd>valuetype_cd0</valuetype_cd>
<tval_char>tval_char0</tval_char>
<nval_num units="units0">3.141592653589</nval_num>
<valueflag_cd name="name2">valueflag_cd0</valueflag_cd>
<quantity_num>3.141592653589</quantity_num>
<units_cd>units_cd0</units_cd>
<end_date>2006-05-04T18:13:51.0Z</end_date>
<location_cd name="name3">location_cd0</location_cd>
</observation>
Is it possible to add an additional column (flag column) in the Observation_Fact table and be able to use the PDO Query Service to return the data row including the added column?
Do I declare this new column in the "i2b2_PDODataType.xsd"?
Thanks,
Johnny Phan
Programmer Analyst
UTHealth
School of Biomedical Informatics
Dat.Q.Phan.uth.tmc
New download sites for the UMass Med School i2b2 Web Client plug-ins & Toolset for the FAIR Initiative, WISE, & ExportXLS v2.0 [01-07-2013]
From: Chan, Wayne [Wayne.Chan.umassmed] Sent: Monday, January 07, 2013 4:42 PM To: 'i2b2 AUG Members' Cc: Sadasivam, Rajani; Houston, Thomas; English, Thomas Subject: RE: New download sites for the UMass Med School i2b2 webclient plugins & Toolset for the FAIR Initiative, WISE, & ExportXLS v2.0
Hi, all,
In response to a request, we have made all our i2b2 tools downloadable kits available from Google Code, in addition to our own UMassMed.edu website. They are as following:
- The FAIR webclient plugins & DBA Toolset:
- The FAIR DBA Toolset –
- From UMassMed.edu (as before) à http://micard.umassmed.edu/fair-dba.html
- From Google Code (new) à http://code.google.com/p/i2b2-fair-dba-toolset/downloads/detail?name=FAIRdbAtools-v1.0-20121113.zip&can=2&q
- Reference wiki (on i2b2 Community Forum wiki, as before) à https://community.i2b2.org/wiki/display/FAIR/FAIR+DBA+Toolset
- The FAIR-viewer plugin –
- From UMassMed.edu (as before) à http://micard.umassmed.edu/fair-viewer.html
- From Google Code (new) à http://code.google.com/p/i2b2-fair-viewer-webclient-plugin/downloads/detail?name=FAIRviewer-v1.0-20121113.zip&can=2&q
- Reference wiki (on i2b2 Community Forum wiki, as before) à https://community.i2b2.org/wiki/display/FAIR/FAIR-viewer
- The FAIR-conceptTracer plugin –
- From UMassMed.edu (as before) à http://micard.umassmed.edu/fair-concept.html
- From Google Code (new) à http://code.google.com/p/i2b2-fair-concept-tracer-webclient-plugin/downloads/detail?name=FAIRconceptTracer-v1.0-20121113.zip&can=2&q
- Reference wiki (on i2b2 Community Forum wiki, as before) à https://community.i2b2.org/wiki/display/FAIR/FAIR-conceptTracer
- The FAIR DBA Toolset –
- The WISE webclient plugins:
- The WISE-annotator –
- From UMassMed.edu (as before) à http://micard.umassmed.edu/wise-annotator.html
- From Google Code (new) à http://code.google.com/p/i2b2-wise-annotator-webclient-plugin/downloads/detail?name=WISEannotator_v1.1_20120320.zip&can=2&q
- Reference wiki (on i2b2 Community Forum wiki, as before) à http://community.i2b2.org/wiki/display/WISE/WISE-annotator
- The WISE-searcher –
- From UMassMed.edu (as before) à http://micard.umassmed.edu/wise-searcher.html
- From Google Code (new) à http://code.google.com/p/i2b2-wise-searcher-webclient-plugin/downloads/detail?name=WISEsearcher_v1.1_20120320.zip&can=2&q
- Reference wiki (on i2b2 Community Forum wiki, as before) à http://community.i2b2.org/wiki/display/WISE/WISE-searcher
- The WISE-annotator –
- The ExportXLS v2.0 (collaboration result with Universita di Pavia):
-
- From UMassMed.edu (as before) à http://micard.umassmed.edu/exportxls.html
- From Google Code (as before, per Mauro) à http://code.google.com/p/i2b2-export-xls-plugin/downloads/detail?name=ExportXLS-v2.0-20120217.zip&can=2&q
- Reference wiki (on i2b2 Community Forum wiki, as before) à https://community.i2b2.org/wiki/display/ExportXLS/ExportXLS+Home
-
Please feel free to contact us regarding any questions or problems. Thanks.
Wayne
S. Wayne Chan, MSEE, MSME
Biomedical Research Informatics Development Group (BRIDG) and Biomedical Research Informatics Consultation & Knowledge Service (BRICKS),
Division of Health Informatics & Implementation Science (HIIS),
Department of Quantitaive Health Sciences (QHS),
University of Massachusetts Medical School (UMMS) at Worcester, MA 01655
(508) 856-8947
From: Chan, Wayne [Wayne.Chan.umassmed] Sent: Monday, December 03, 2012 10:09 AM To: 'i2b2 AUG Members' Cc: Sadasivam, Rajani; Houston, Thomas; English, Thomas Subject: The UMass Med School i2b2 webclient plugins & DBA Toolset for the FAIR Initiative
Hi, all,
We at the University of Massachusetts Medical School are happy to share with the rest of the i2b2 community the FAIR (Familial, Associational, and Incidental Relationships) i2b2 webclient plugins and DBA Toolset that were announced in the recent CTSA iKFC Meeting. They are available for download in our corresponding website page, http://micard.umassmed.edu/software.html (near the bottom).
We have also set up corresponding i2b2 Community project Wiki pages (https://community.i2b2.org/wiki/display/FAIR/Home) about these new tools. This is the link at the dashboard page:
Please don't hesitate to contact us concerning any questions or problems. Thanks.
Wayne
S. Wayne Chan, MSEE, MSME
Biomedical Research Informatics Development Group (BRIDG) and Biomedical Research Informatics Consultation & Knowledge Service (BRICKS),
Division of Health Informatics & Implementation Science (HIIS),
Department of Quantitaive Health Sciences (QHS),
University of Massachusetts Medical School (UMMS) at Worcester, MA 01655
(508) 856-8947
Handling patient class (in, out, emergency, etc.) [01-03-2013]
From: Lee, Eunjung S [sallylee.u.washington] Sent: Thursday, January 03, 2013 7:21 PM To: Murphy, Shawn N.; Harle,Christopher A; i2b2 AUG Members Subject: RE: Handling patient class (in, out, emergency, etc.)
Thanks all for the great feedback!
We will create the Visit Type ontology as well (although a very small and short one).
Sally
E. Sally Lee, Ph.D
Biomedical Informatics Core Consult
Amalga Clinical Data Repository
Institute of Translational Health Sciences
University of Washington
sallylee.uw
From: Murphy, Shawn N. [SNMURPHY.PARTNERS] Sent: Thursday, January 03, 2013 9:07 AM To: Harle,Christopher A; Lee, Eunjung S; i2b2 AUG Members Subject: RE: Handling patient class (in, out, emergency, etc.)
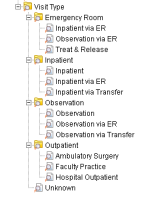
Very nice visit-type layout Chris. Also, Sally, if you are looking to do queries like "was this diagnosis made during an outpatient encounter" you will want to use the {}Selected Groups Occur in the same Financial Encounter{} option when you do the query with the diagnosis in one panel and the "outpatient" item in the other group.
Thanks,
Shawn.
From: Harle,Christopher A [charle.phhp.ufl] Sent: Thursday, January 03, 2013 11:05 AM To: 'Lee, Eunjung S'; i2b2 AUG Members Subject: RE: Handling patient class (in, out, emergency, etc.)
Our approach has been the following.
Hope this helps. --Chris
Christopher Harle, PhD
Assistant Professor
Department of Health Services Research, Management and Policy
College of Public Health and Health Professions
University of Florida
phone: 352-273-6081
e-mail: charle.phhp.ufl
From: Lee, Eunjung S [sallylee.u.washington] Sent: Wednesday, January 02, 2013 4:38 PM To: i2b2 AUG Members Subject: Handling patient class (in, out, emergency, etc.)
Hello,
We are trying to figure out how to handle patient class such as inpatient, outpatient, emergency patient, etc. in i2b2 and wanted to ask how i2b2 community is implementing this?
We have visit data with this information, but not sure where or how to put information into the ontology so that people can easily query information such as, "outpatient who has certain diagnosis".
Thank you for your help!
Yours,
Sally
E. Sally Lee, Ph.D
Biomedical Informatics Core Consult
Amalga Clinical Data Repository
Institute of Translational Health Sciences
University of Washington
sallylee.uw