Genomics ontology presents the genetic variants as query-able concepts in the i2b2 web-client. Creating this queryable ontology is a 3-step process:
- Create Concepts in Concept_Dimension table
- Create entries in metadata table (metadata_genomics in our example)
- Create an entry in the Table_Access table
Concepts Entries:
Variants are differences between two genomes. We have considered the following 2 important types of variants:
- SNP
- INDEL
Each of these variants can be queried by rs identifier or by gene name, thus giving a total of 4 concepts. Sample scripts to insert theses 4 concepts in the concept_dimension table can be found inside the package in “Scripts for Sample Data” folder.
Concept codes that has been used in the Sample Data are as follows
Variant/Concept Name | Concept Code |
SNP | SO:0001483 |
indel | SO:1000032 |
Metadata Entries:
The metadata table entries determine the Ontology hierarchical structure of concepts as presented by i2b2 web-client. They also determine the names and data type of concepts and operators to apply on them as part of ValueMetadata in the “C_METADATAXML” field in metadata table (metadata_genomics in our example)
- Value metadata
There are two value metadata types that accompany the two variant concepts, SNP and indel. The SNP variant concept has the datatype = GENETIC_VARIANT_SNP and indel variant concept has the datatype = GENETIC_VARIANT_INDEL. The concept code of each type is contained in the TestID tag.
<?xml version="1.0"?>
<ValueMetadata>
<Version>3.03</Version>
<CreationDateTime>01/28/2016</CreationDateTime>
<TestID>SO:0001483</TestID>
<TestName>SNP</TestName>
<DataType>GENETIC_VARIANT_SNP</DataType>
<Oktousevalues />
<MaxStringLength>30</MaxStringLength>
<EnumValues />
<UnitValues>
<NormalUnits/>
</UnitValues>
</ValueMetadata>
<?xml version="1.0"?>
<ValueMetadata>
<Version>3.03</Version>
<CreationDateTime>01/28/2016</CreationDateTime>
<TestID>SO:1000032</TestID>
<TestName>indel</TestName>
<DataType>GENETIC_VARIANT_INDEL</DataType>
<Oktousevalues />
<MaxStringLength>30</MaxStringLength>
<EnumValues />
<UnitValues>
<NormalUnits/>
</UnitValues>
</ValueMetadata>
Sample scripts to create and insert Genomics data in metadata_genomics table can be found inside the package in “Scripts for Sample Data” folder.
Table_Access Entries:
Table_Access table entry determines which metadata table should i2b2 consider to generate the correct Ontology structure.
Sample scripts to insert Genomics Table_Access data in can be found inside the package in “Scripts for Sample Data” folder.
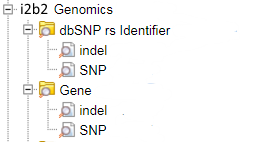
Once all the scripts are executed to successfully load the Ontology in the database the following ontology structure can be found in Ontology section in the i2b2 web-client:
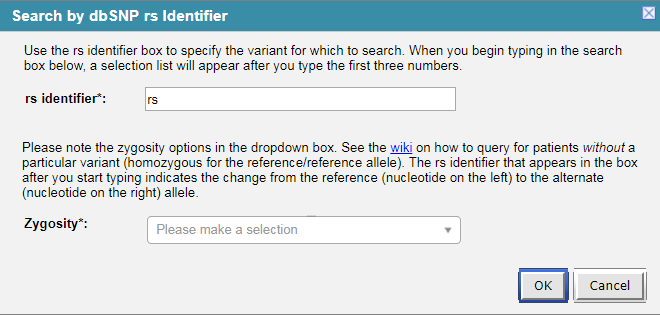
- To query this ontology a user will drag any of the above concepts from the “Navigate Terms” into the “Query Tool” area and a dialog box will prompt the user to enter the text by which they want to search the blob field.
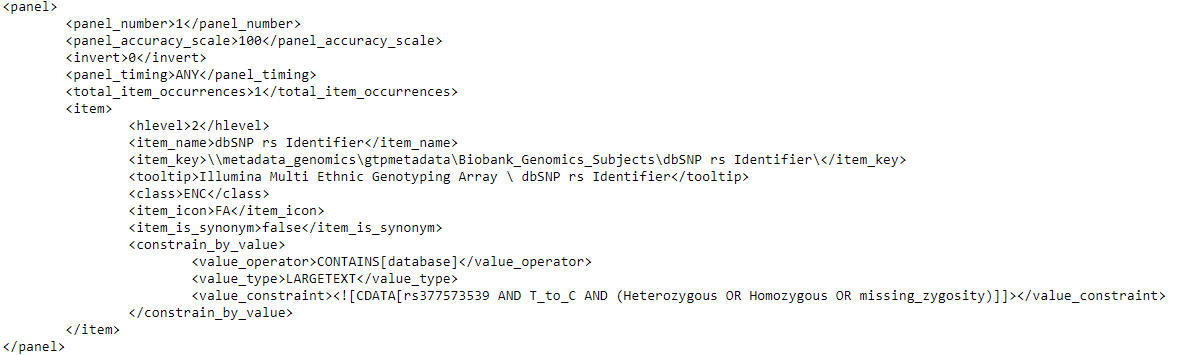
- When a user will fill in the input fields (with rs_identifier = “rs377573539 | T to C” and Zygosity = “Select all” for example) and run the query, the following query request XML will be generated by the Web Client:
- In the background the CRC will convert the request XML to a proper SQL Server contains statement (in this example below):
with t as (
select f.patient_num
from dbo.observation_fact f
where
f.concept_cd IN
(
select concept_cd
from dbo.concept_dimension
where concept_cd IN ('SO:0001483','SO:1000032')
)
AND (modifier_cd = '@' AND valtype_cd = 'B'
AND CONTAINS(observation_blob,'rs377573539 AND T_to_C AND (Heterozygous OR Homozygous OR missing_zygosity)')
)
After the query is executed all patients matching the input genomic variants will be returned to the i2b2 web client.