- Dashboard
- Health Ontology Mapper
- HOM Home
- Attachments
- AMIA_Spring_IDR.doc
AMIA_Spring_IDR.doc
The Integrated Data Repository:
A Non-Traditional Data Warehousing Approach for the Translational Investigator
Marco J. Casale 3 , MS, Russ J. Cucina 1 , MD, MS, Mark G. Weiner 2 , MD, David A. Krusch, MD 3, Prakash Lakshminarayanan 1 , MBA, Aaron Abend, Bill Grant, Rob Wynden 1 , BSCS
1 University of California, San Francisco, CA; 2 University of Pennsylvania, Philadelphia, PA;
3 University of Rochester, Rochester, NY
Abstract
An integrated data repository (IDR) containing aggregations of clinical, biomedical, economic, administrative, and public health data are key components of an overall translational research infrastructure. But most available data repositories are designed using standard data warehouse architecture using a predefined data model, which does not facilitate many types of health research. In response to these shortcomings we have designed a schema and associated components which will facilitate the creation of an IDR by directly addressing the urgent need for terminology and ontology mapping in biomedical and translational sciences and give biomedical researchers the required tools to streamline and optimize their research. The proposed system will dramatically lower the barrier to IDR development at biomedical research institutions to support biomedical and translational research, and will furthermore promote inter-institute data sharing and research collaboration.
Introduction
An integrated data repository (IDR) containing aggregations of clinical, biomedical, economic, administrative, and public health data are key components of an overall translational research infrastructure. Such a repository can provide a rich platform for a wide variety of biomedical research efforts. Examples might include correlative studies seeking to link clinical observations with molecular data, data mining to discover unexpected relationships, and support for clinical trial development through hypothesis testing, cohort scanning and recruitment. Significant challenges to the successful construction of a repository exist, and they include, among others, the ability to gain regular access to source clinical systems and the preservation of semantics across systems during the aggregation process.
Most repositories are designed using standard data warehouse architecture, with a predefined data model incorporated into the database schema. The traditional approach to data warehouse construction is to heavily reorganize and frequently to modify source data in an attempt to represent that information within a single database schema. This information technology perspective on data warehouse design is not well suited for the construction of data warehouses to support translational biomedical science. The purpose of this paper is to discuss the components which would facilitate the creation of an IDR by directly addressing the need for terminology and ontology mapping in biomedical and translational sciences and the novel approach to data warehousing design.
Background
The IDR database design is a departure from the traditional data warehouse design proposed by Inmon. The data warehouse architecture model shown in Figure 1 depicts the process of transforming operational data into information for the purpose of generating knowledge. The diagram displays data flowing from left to right in accordance with the corporate information factory (CIF) approach (Inmon et al). According to Inmon, the data enters the CIF as raw data collected by operational applications. The data is transformed through extract, transform and load processes and stored in either the data warehouse or the ODS, operational data store.
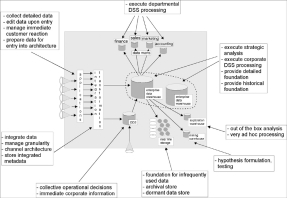
Figure 1. Inmon’s Corporate Information Factory Data Warehousing Model
“Often up to 80 percent of the work in building a data warehouse is devoted to the extraction, transformation, and load (ETL) process: locating the data; writing programs to extract, filter, and cleanse the data; transforming it into a common encoding scheme; and loading it into the data warehouse.” (Hobbs, Hillson & Lawande,) The data model is typically designed based on the structure of the source data. Figure 2 depicts a source system clinical data model.
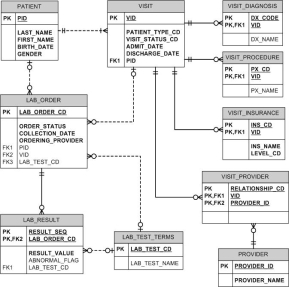
Figure 2. University of Rochester simplified clinical data repository OLTP data model
Furthermore, after the data has been prepared, it is loaded into the de-normalized schema of the data warehouse or data marts and resides there in a fine grain level of detail. The logical design of a data warehouse is usually composed of the star schema. “A star schema is a simple database design (particularly suited to ad-hoc queries) in which dimensional data (describing how data are commonly aggregated) are separated from fact or event data ( describing individual business transactions).” (Hoffer, Prescott & McFadden)
However, an IDR has to incorporate heterogeneous data. A common data modeling design approach in biomedical informatics is the entity-attribute-value (EAV) model. An EAV design , conceptually involves a table with three columns—a column for entity/object identification (ID), one for attribute/parameter (or an attribute ID pointing to an attribute descriptions table), and one for the value for the attribute. The table has one row for each A-V pair. The IDR is built on the combination of both the star schema data model and the EAV approach. An example of this approach can be seen in Partners Healthcare i2b2 NIH supported data warehouse design.
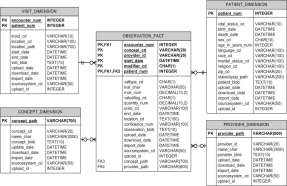
Figure 3. Partners Healthcare i2b2 star schema EAV data model
EAV design is potentially attractive for databases with complex and constantly changing schemas that reflect rapidly evolving knowledge in scientific domains. Here, when a conventional design is used, the tables (and the code that manipulates them, plus the user interface) need continuous redesign with each schema revision.
Discussion
There are several challenges posed by IDR projects geared toward biomedical research which do not apply to the construction of most commercial warehouse implementations: 1) integrity of source data - a clear requirement in the construction of an IDR is that source data may never be altered, nor may their interpretation be altered. Records may be updated, but strict version control is required to enable reconstruction of the data that was available at a given point in time. Regulatory requirements and researchers demand clear visibility to the source data in its native format to verify it has not been altered; 2) high variability in source schema designs - IDRs import data from a very large set of unique software environments, from multiple institutions, each with its own unique schema; 3) limited resources for the data governance of standardization - widespread agreement on the interpretation, mapping and standardization of source data that has been encoded using many different ontologies over a long period of time may be infeasible. In some cases the owners of the data may not even be available to work on data standardization projects, particularly in the case of historical data; 4) limited availability of software engineering staff with specialized skill sets - interpretation of source data during the data import process requires a large and highly skilled technical staff with domain expertise, talent often not available or only at considerable expense; and 5) valid yet contradictory representations of data - there are valid, yet contradictory interpretations of source data depending on the domain of discourse of the researcher. Examples related to the inconsistency of the researchers’ domain of discourse include: two organizations may interpret the same privacy code differently, or researchers within the same specialty may not use the same ontology, or clinical and research databases often encode race and ethnicity in differing ways. We have developed an alternative approach that incorporates the use of expert systems technologies to provide researchers with data models based on their own preferences, including the ability to select a preferred coding/terminology standard if so desired. We believe that such an approach will be more consistent with typical research methodologies, and that it will allow investigators to handle the raw data of the repository with the degrees of freedom to which they are accustomed.
An ontology mapping component provides a s treamline data acquisition and identification process by delivering data to researchers in a just-in-time fashion, instead of requiring that all data be transmitted to the IDR via a single common format and without the requirement that all data be stored within a single centralized database schema, providing a knowledge management system and ontology mapping tools that enable less technical users to translate the complex array of data fields needed to fulfill data requests, and facilitating inter-institutional data sharing by translating data definitions among one or more site-specific terminologies or ontologies, and shareable aggregated data sets.
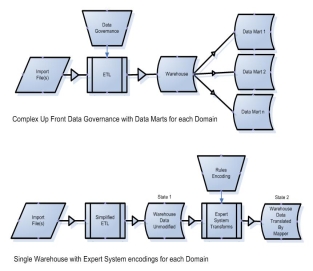
Figure 2. Complex data governance (top) can be exchanged for rules encoding (bottom)
2) Translation - the translation of data from its source ontology into the ontology required by the researcher will not be completed during the extract, transform and load (ETL) phase. The ontology mapping will be completed after the source data has already been imported into the IDR.
To support the translation of data, we are developing an approach called Inference Based Ontology Mapping – in which the source data must be translated into the ontology that the biomedical researcher requires for a particular domain of expertise. The IDR will use a rules-based system to perform this mapping of source data format to the researcher’s ontology of choice.
Federated vs. Centralized Approach
The debate regarding a federated data warehouse design versus an ‘all in one’ centralized data warehouse design has been a key point between the two main data warehousing factions between Bill Inmon and Ralph Kimbal. The following example outlines the differences for the two approaches and justifies why the centralized approach is the favorable choice in biomedical informatics .
Many institutions have electronic clinical data that is decentralized across different departments. That infrastructure can be used to create an integrated data set with information that spans the source systems. However, the decentralization creates the need for redundant steps, lengthy iterative processes to refine the information, and requires that more people have access to protected health information in order to satisfy the research information needs. To illustrate these issues, the following describes the workflow needed to define a cohort of people known to have cardiovascular disease and laboratory evidence of significant renal disease defined by an elevated serum creatinine.
In the decentralized system, where should the investigator start? He can begin by going to the billing system that stores diagnoses and get a list of PHI (Personal Health Information) of people with a history of a heart attack. Then he can transport that list of identifiers to the people who work in the laboratory and request the serum creatinine levels on that set of patients, and then limit the list to those who have an elevation. The lab will have to validate the patient matches generated by the billing system by comparing PHI, a step redundant with the billing system. Furthermore, many of the subjects associated with heart attack may not have the elevated creatinine, so, in retrospect, the PHI of these people should not have been available to the people running the query in the lab. Perhaps the cohort that was generated was not as large as expected, and the investigator decides to expand the cohort to those patients with a diagnosis of peripheral vascular disease and stroke. He then has to iterate back to the billing system to draw additional people with these additional diagnoses, and then bring the new list of patient identifiers to the lab to explore their creatinine levels.
The centralized warehouse as proposed will conduct the matching of patient identifiers behind the scenes. The information system will conduct the matching of patients across the different components of the database, so that identifiers do not have to be manually transported and manipulated by the distinct database managers at each location. The result is that a centralized warehouse is radically more secure than a decentralized warehouse due to the reduced exposure of PHI. Further, if the query produces results that are not satisfactory, the cycle of re-querying the data with new criteria will be faster, and user controlled.
Conclusion
Our proposed design is intended to greatly facilitate biomedical research by minimizing the initial investment that is typically required to resolve semantic incongruities that arise when merging data from disparate sources. Through the use of a rules-based system, the translation of data into the domain of a specific researcher can be accomplished more quickly and efficiently than with a traditional data warehouse design. The proposed system will dramatically lower the barrier to IDR development at biomedical research institutions to support biomedical and translational research, and promote inter-institute data sharing and research collaboration.
References
- Brinkley JF, Suciu D, Detwiler LT Gennari JH, Rosse C. A framework for using reference ontologies as a foundation for the semantic web. Proc. AMIA Symp. 2006; 96-100.
- Gennari JH, Musen MA, Fergerson RW, Grosso WE, Crubézy M, Eriksson H, Noy NF, Tu SW. The evolution of Protégé: an environment for knowledge-based systems development. International Journal of Human Computer Studies 2003; 58(1):89-123.
- Advani A, Tu S, O’Connor M, Coleman R, Goldstein MK, Musen M. Integrating a modern knowledge-based system architecture with a Legacy VA database: The ATHENA and EON projects at Stanford. Proc. AMIA Symp. 1999; 653-7.
- Hobbs, Lilian; Hillson, Susan & Lawande,
Shilpa Oracle9iR2 Data Warehousing. Burlington: Digital Press. 2003, 6
- Inmon, W. H., Imhoff, C., Sousa, R. Introducing
the Corporate Information Factory 2nd Edition New York: John Wiley & Sons, Inc. 2001
- Hoffer, Jeffrey A.; Prescott, Mary B., McFadden,
Fred R.. Modern Database Management. Upper Saddle River: Prentice Hall. 2002, 421
- Nadkarni, Prakash M. MD et al,
Organization of Heterogeneous Scientific Data Using the EAV /CR Representation . Journal of American Medical Informatics 1999
Goals:
-
Biomedical Informatics ontologies require a unique approach to the standard data warehousing design
- Use of the EAV data model
-
There is a necessity to map from an informal or unstructured vocabulary to a formal ontology. This approach requires a database design which captures the map, the encoding of the source term and destination term.
- Remove heavy lifting or large effort of metadata cross mapping of terms during the ETL process. The terms can be mapped as new requests from translational scientists present requests.
- Allows the translational researcher to data mine the IDR in their chosen ontology
- The onotolgy map is created at the point of request instead of at the point of ETL
- Maps can be shared amongst CTSA universities