REDCap-to-i2b2 Loader
James Gregoric
March 7, 2012
Concept
The REDCap-to-i2b2 Loader is a web-based application that facilitates the porting of REDCap study data to i2b2. The ported data consists of study metadata (including a study ontology) and the study's clinical data.
User Operation
Execution begins with the login screen, which uses table-based authorization:
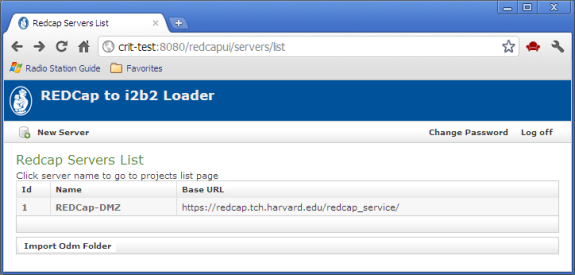
After login, a list of available REDCap servers is displayed:
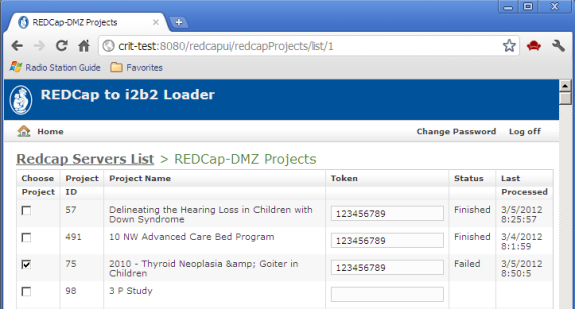
Selecting a server, a list of REDCap projects on the selected server is displayed:
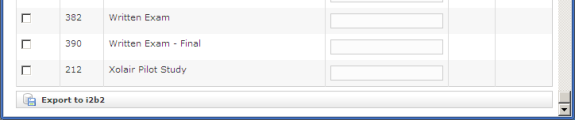
Simply check the projects to be exported to i2b2 and press the 'Export to i2b2' button. The time required for a "typical" project (say 100 fields by 100 subjects) will be on the order of two to five minutes.
Notes:
- Token access to selected projects is required. A project's token may be obtained from the administrator for the REDCap instance.
- The Last Processed timestamp is displayed for projects that have already been exported, along with the status of the export attempt.
- If a project that has already be exported is re-selected for export, the export process begins by cleaning out all references to the project from the i2b2 database. That is, a REDCap project that is imported into i2b2 more than once will contain only records from the most recent successful import; all traces of previous exports will be gone.
i2b2 UI
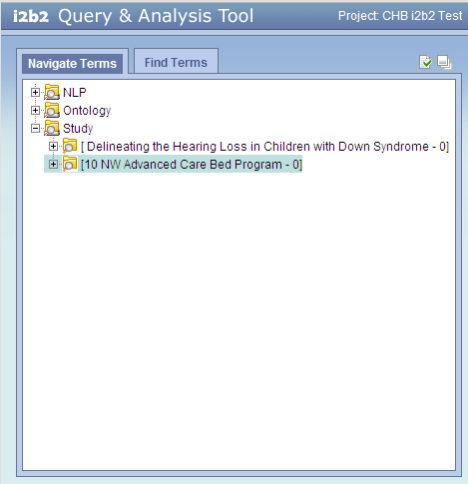
The i2b2 screen shot below shows the ontology folders for two REDCap studies.
Notice that:
- The ontology for REDCap studies appears in the 'Study' folder, separate from the 'Ontology' folder.
- Each REDCap study produces its own ontology, seen in i2b2 as a folder within the Study folder.
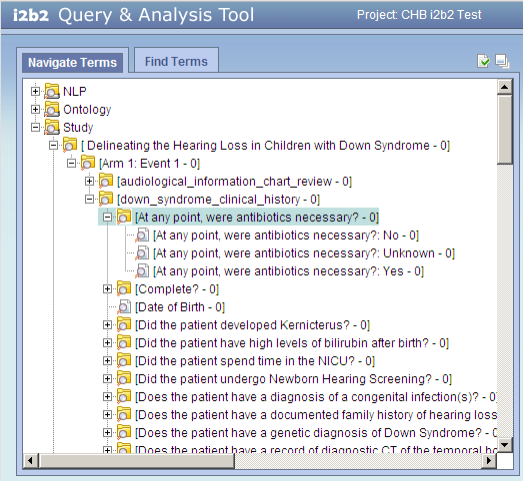
The i2b2 screen shot below shows the expanded ontology folders for the first REDCap study (Delineating the Hearing Loss...). The question "At any point, were antibiotics necessary?" has three possible responses, each a separate entry in the ontology. This question appears in the form named "down_syndrome_clinical_history", which in turn appears in "Arm 1: Event 1", which is the first arm-event in the study.
Notice that:
- The ontology for a study is a hierarchy of folders:
arm-event*
form
question
possible responses
*arm and event are concatenated because the i2b2 ontology structure does not include the concept of a study arm.
- Each ontology entry (leaf node) is a possible response for a particular
arm-event/form/question.
Process Overview
The process proceeds in several steps:
The first step extracts the selected study metadata and clinical data from the REDCap database and translates it into ODM XML, which is saved as a file in an archive directory. The archive directory serves as a common point of exchange - any process can drop an ODM XML file into this directory, to be picked up by subsequent steps in the porting process to end up in the i2b2 production database.
The ODM XML comprises two main sections - the metadata and the clinical data:
<?xml version="1.0" encoding="UTF-8" standalone="yes" ?>
<ODM ODMVersion="1.3.1"
CreationDateTime="2012-02-03T10:59:14.175-05:00"
FileOID="000-000-000"
FileType="Transactional"
xmlns:ns2=http://www.w3.org/2000/09/xmldsig#
xmlns="http://www.cdisc.org/ns/odm/v1.3">
<Study OID="10">
<GlobalVariables>
</GlobalVariables>
<BasicDefinitions />
<MetaDataVersion Name="Version 1.3.1" OID="v1.3.1">
.
.
.
</MetaDataVersion>
</Study>
<ClinicalData MetaDataVersionOID="v1.3.1" StudyOID="10">
.
.
.
</ClinicalData>
</ODM>
The second step brings the ODM XML into an i2b2 staging schema. This schema is without indexes, thus improving insertion performance, as well as providing a means to vet the data before it makes its way into the production database.
The final step consists of an ETL process to bring the data into the production database.