Last Updated: 03/18/2013
i2b2 Timeline performance with 800M+ facts [09-26-2012]
From: Dan Connolly [dconnolly.kumc] Sent: Wednesday, September 26, 2012 3:03 PM To: i2b2 AUG Members Subject: i2b2 timeline performance with 800M+ facts?
Performance of the i2b2 timeline was OK (a few seconds latency) until our late August release, when our fact count went over 800M. Since then, PDO queries take orders of magnitude longer (hundreds of seconds, if it finshes at all). Does anyone else use the timeline on their whole repository? Or do you only use it on smaller projects? How is the performance? We're using bitmapped indexes (a suggestion we picked up from Peter Beninato). They work great, as long as Oracle uses both the patient_num and the concept_cd index. But for reasons we can't figure out, it sometimes Oracle does something else. Does anyone have suggestions on how to diagnose, let alone fix, this sort of performance problem?
Dan Connolly, KUMC Medical Informatics
i2b2 Tutorial Nov. 7, 2012 [09-20-2012]
From: Daniele Segagni [daniele.segagni.fsm.it] Sent: Thursday, September 20, 2012 3:02 AM To: Churchill, Susanne E. Cc: i2b2 members Subject: Re: i2b2 Tutorial Nov. 7, 2012
Dear Susanne, is possible to follow this interesting tutorial in a remote manner, for example using WebEx or something similar? Thank you, regards Daniele
Ing. Daniele Segagni
Laboratorio di Informatica e Sistemistica per la Ricerca Clinica
IRCCS Fondazione S.Maugeri, Pavia, Italy
tel: +39 0382 592038
Churchill, Susanne E. ha scritto:
Dear Colleagues,
Mike Mendis will be offering a three hour i2b2 tutorial on Wednesday evening, November 7 (6-9 pm) (description below). This is the evening before the actual CTSA KFC Meeting and on the last day of the AMIA Symposium. Note this will take place at Northwestern (we're waiting for a specific location) NOT the AMIA hotel.
Please register with me if you would like to participate.
Thanks,
Susanne
Susanne E. Churchill, Ph.D.
Executive Director
i2b2/Nat'l Center for Biomedical Computing
77 Avenue Louis Pasteur, Room 255
Boston, MA 02115
tel: (617) 525-4465
fax: (617 525-4491
HOW TO "i2b2" TUTORIAL DESCRIPTION
We will describe a step by step approach to implementing i2b2 at your institution, what technologies are used and any third party software that is required, such as application or database servers. We will continue by writing a new analytic "cell" and leveraging advanced services such as natural language processing. The new cell will be a fully working i2b2 cell that can receive and respond to i2b2 calls. Once the cell has been developed, an i2b2 webclient plugin will be developed that sends a message to the cell and displays the response. We conclude with a roadmap for the near term and longer term i2b2 developments.
Due to the complex nature of this tutorial, the participants should download and install the tutorial Virtual Machine (VM; see below) prior to attending. The VM will run on any Windows or Macintosh machine and will contain a complete and installed i2b2 server, i2b2 webclient, application server, database server and any developments needed. A completed cell and plugin will also be included.
Prerequisites: The participant should download and install the source code of the "i2b2 Tutorial" that is available at https://www.i2b2.org/software/ctsa2012.html prior to arriving at the workshop.
Intended Audience: Scientists; researchers; healthcare analysts, database programmers, and other healthcare professionals; academic faculty or professionals setting up bioinformatics facilities and/or relating these to clinical data repositories and computer scientists, system developers, and programmers.
Content level: 20% Basic, 50% Intermediate, 30% Advanced.
Proper usage of modifier folders versus containers [09-07-2012]
From: Nathan Graham [ngraham.kumc] Sent: Friday, September 07, 2012 1:33 PM To: 'Nathan Graham'; Phillips, Lori C.; Mendis, Michael E. Cc: i2b2 AUG Members Subject: RE: Proper Usage of Modifier Folders versus Containers
Lori,
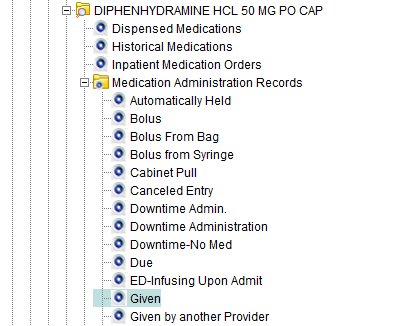
Screenshot attached:
Nathan Graham
Software Engineer
Department of Biostatistics
University of Kansas Medical Center
ngraham.kumchttp://informatics.kumc.edu
"Phillips, Lori C." <LCPHILLIPS.PARTNERS> 9/7/2012 11:51 AM
Can you send a screen shot of the query tool after the modifier is dropped into it?
From: Nathan Graham [ngraham.kumc] Sent: Friday, September 07, 2012 12:44 PM To: 'Nathan Graham'; Phillips, Lori C.; Mendis, Michael E. Cc: i2b2 AUG Members Subject: RE: Proper Usage of Modifier Folders versus Containers
Lori,
Thanks for the quick response!
This query was from the web client (version 1.6.03). I've attached the incoming/outgoing XML - it looks like it's referencing the modifier and the modifier folder - I don't see the concept code in there. The query was run against one of our medications in the ontology (see attached screen shot). Generated SQL from this query is earlier in the thread (double-checked that it matches the XML).
Thanks again!
Regards,
Nathan
Nathan Graham
Software Engineer
Department of Biostatistics
University of Kansas Medical Center
ngraham.kumchttp://informatics.kumc.edu
"Phillips, Lori C." <LCPHILLIPS.PARTNERS> 9/7/2012 9:30 AM
Nathan, I ran your case on our system using the workbench and it ran fine.I was using hive version 1.6.05 and used the following modifier folder term from the Demo set in my query:Diagnoses/Neoplasms/Malignant neoplasms/TNM Staging/TMN Staging, Distant Metastisis/
1. What version of the hive are you using and are you running on the webclient or the workbench?
2. When you run the query what does the outgoing query tool request tool like?
In particular I am wondering what the <panel> xml looks like..
<panel>
<panel_number>1</panel_number>
...
<item>
<hlevel>3</hlevel>
<item_name>Malignant neoplasms</item_name>
<item_key>\\i2b2_DIAG\i2b2\Diagnoses\Neoplasms (140-239)\Malignant neoplasms (140-208)\</item_key>...
<constrain_by_modifier>
<modifier_name>TNM Staging, Distant Metastisis</modifier_name>
<modifier_key>\\i2b2_DIAG\TNM\Metastisis\</modifier_key>
<applied_path>\i2b2\Diagnoses\Neoplasms (140-239)\Malignant neoplasms (140-208)%</applied_path>
</constrain_by_modifier>
You should see both the modifier and the concept it applies to in the <panel> xml..
Lori
From: Nathan Graham [njgraham.gmail] Sent: Friday, September 07, 2012 9:58 AM
To: Mendis, Michael E.; Phillips, Lori C.Subject: Proper Usage of Modifier Folders versus Containers
Mike/Lori, I apologize if this e-mail is a duplicate - our mail servers at Kansas University Medical Center keep reporting that they can't find "partners.org". So, I thought I'd send this from my GMail account for now until I figure out why our mail servers don't seem to like the Partners address. I'm trying to use modifier folders (visual_attribute:'DA') and it appears as if the generated sql for a modifier inside a folder doesn't include the parent concept_cd so we don't get the right query result.However, when we use a modifier container (visual_attribute:'OA') the generated sql does include the concept_cd and therefore appears to generate the correct result. I've included my e-mail to the AUG below which has generated SQL from i2b2.
It appears to be a bug - can you repeat this behavior? Or, can yousee what I'm doing wrong? I searched the i2b2 jira site but didn't find anything on this topic.
Thanks. Regards,
Nathan
Nathan GrahamSoftware EngineerDepartment of BiostatisticsUniversity of Kansas Medical Center ngraham.kumc http://informatics.kumc.edu
Nathan Graham 9/4/2012 5:19 PM
AUG members, I'm looking for some advice on how to properly use modifier foldersand containers.
When I tried to use modifier folders(visual_attribute:'DA'), the generated query for a modifier inside thefolder doesn't include the parent concept code but includes the parentmodifier folder instead. This query does not yield correct results.
When I then tried modifier containers instead (visual_attribute:'OA'),the query did include the parent concept (and, yielded what I believeto be correct results). I would expect that the folders and containers would work the same way as far as the generated SQL.
Has anyone successfully used modifierfolders? If so, any suggestions? I've included generated SQL below.
We're using i2b2 1.6.03. using "DA" (parent concept not referenced)
INSERT INTO BlueHerondata.QUERY_GLOBAL_TEMP ( patient_num , panel_count )SELECT patient_num , 1 AS panel_countFROM (SELECT /+ index(observation_fact fact_cnpt_pat_enct_idx) */ patient_num FROM BlueHerondata.observation_fact WHERE MODIFIER_CD IN (SELECT MODIFIER_CD FROM BlueHerondata.MODIFIER_DIMENSION WHERE MODIFIER_PATH LIKE '\Medication\MAR%' --This is the modifier folder ) AND ( (MODIFIER_CD IN (SELECT MODIFIER_CD FROM BlueHerondata.MODIFIER_DIMENSION WHERE MODIFIER_PATH LIKE '\Medication\MAR\Given%' --This is thespecific modifier inside the MAR folder noted above )) ) GROUP BY patient_num ) t ;INSERT INTO BlueHerondata.DX ( patient_num )SELECT *FROM ( SELECT DISTINCT patient_num FROM BlueHerondata.QUERY_GLOBAL_TEMP WHERE panel_count = 1 ) q using "OA" - parent concept is now referenced. INSERT INTO BlueHerondata.QUERY_GLOBAL_TEMP ( patient_num , panel_count ) SELECT patient_num , 1 AS panel_countFROM (SELECT /+ index(observation_fact fact_cnpt_pat_enct_idx) */ patient_num FROM BlueHerondata.observation_fact WHERE concept_cd IN (SELECT concept_cd FROM BlueHerondata.concept_dimension WHERE concept_path LIKE '\i2b2\Medications%' --The appropriateparent concept for the modifier ) AND ( (MODIFIER_CD IN (SELECT MODIFIER_CD FROM BlueHerondata.MODIFIER_DIMENSION WHERE MODIFIER_PATH LIKE '\Medication\MAR\Given%' --The modifierwe selected )) ) GROUP BY patient_num ) t ;INSERT INTO BlueHerondata.DX ( patient_num )SELECT *FROM ( SELECT DISTINCT patient_num FROM BlueHerondata.QUERY_GLOBAL_TEMP WHERE panel_count = 1 ) q;
Thanks!
Regards,
Nathan
Nathan GrahamSoftware EngineerDepartment of BiostatisticsUniversity of Kansas Medical Center ngraham.kumc http://informatics.kumc.edu
Claims database and i2b2 [09-07-2012]
From: Valtchinov, Vladimir I.,Ph.D. Sent: Friday, September 07, 2012 1:16 PM To: 'Lake, Amy K.'; i2b2 AUG Members Subject: RE: Claims database and i2b2
Hi Amy-
I am working on an i2b2 implementation for the CMS MID project
(http://www.healthcareitnews.com/news/cms-selects-5-participants-medicare-imaging-demonstration-project) at the Brigham radiology end.
It is about quantifying appropriateness of medical imaging ordering, and as such has medical claims data as one of its components. We are almost there with the first phase, the pure CPOE data for the Brigham being the convener, with the next data source being claims for the imaging exams.
There are some obvious and also some hidden pitfalls in using the patient-centric i2b2 Star Schema for modeling such data, but in the end the schema is sufficient and the major additions are the ontologies. The CPT4's (and possibly the internal exam codes) are good examples of that.
Let me know if you'd like to chat further about our experience, or with more specific questions.
Thanks,
Vlad Valtchinov
CEBI
From: Russ Waitman [rwaitman.kumc] Sent: Friday, September 07, 2012 12:04 PM To: Amy K. Lake; i2b2 AUG Members Cc: darren.henderson.uky; todd.r.johnson.uky Subject: Re: Claims database and i2b2
Hi Amy,
I'd check with the people at the University of Kentucky (possibly Todd Johnson or Darren Henderson) as I think that's a major focus of their i2b2 system. We have more clinical, research, and billing data in our repository,
Russ Waitman
Associate Professor, Director of Medical Informatics
Department of Biostatistics
Assistant Vice Chancellor for Enterprise Analytics
University of Kansas Medical Center
913-945-7087
rwaitman.kumchttp://informatics.kumc.edu
From: Lake, Amy K. [lakea.health.missouri] Sent: Friday, September 07, 2012 11:02 AM To: i2b2 AUG Members Subject: Claims database and i2b2
Hello all, we are new to this listserv. And, we are considering adopting i2b2 for our Missouri State Medicaid claims database. This database includes several years worth of data, millions of individual records with costs on all transactions/services that were billed by health care providers to our State Medicaid program, along personal identifiers, ICDs, CPTs, etc. No clinical information is included. So, we can see if a procedure was provided but not the outcome. For example, we can see if a blood pressure was checked but not what that person's actual blood pressure reading was. We use it extensively for research and statewide policy analysis and hope that adopting i2b2 will facilitate both.
Is anyone else using i2b2 with a medical claims database or a non-clinical database? If so, we would love to get some guidance before we dive in. Please contact me if you have any expertise or ideas about this. Thanks much.
Amy Lake
MO HealthNet Data Project Director
Center for Health Policy
University of Missouri-Columbia
215 Middlebush
Columbia MO 65211
573-882-5412
lakea.missouri
1.6.06 Released [09-07-2012]
From: Murphy, Shawn N. Sent: Friday, September 07, 2012 10:36 AM To: i2b2 AUG Members Subject: 1.6.06 is released
Some minor web client enhancements and a fix to Ontology cell so it doesn't serve Hidden ontology items by default.
Thanks,
Shawn.