Last Updated: 03/19/2013
HIPAA Safe Harbor and implications on age over 90 in i2b2 [12-31-2012]
 From: Russ Waitman [rwaitman.kumc] Sent: Monday, December 31, 2012 2:12 PMTo: Malin, Bradley A; i2b2 AUG Members; Nathan GrahamSubject: RE: HIPAA Safe Harbor and implications on age over 90 in i2b2
From: Russ Waitman [rwaitman.kumc] Sent: Monday, December 31, 2012 2:12 PMTo: Malin, Bradley A; i2b2 AUG Members; Nathan GrahamSubject: RE: HIPAA Safe Harbor and implications on age over 90 in i2b2
Hi Brad,
Thanks for the reply and it's right in line with what I was asking (complying with Safe Harbor).
So it sounds like as we're thinking we need to address events in the past not just the birthdate along the lines of your comments under #3 if we want to strive to be de-identified.
Russ
From: Russ Waitman [rwaitman.kumc] Sent: Monday, December 31, 2012 12:36 PMTo: Murphy, Shawn N.; Mendis, Michael E.Cc: Luke Rasmussen; b.malin.vanderbilt; i2b2 AUG Members; Nathan Graham; Andrew Q Winter; elliotts.amazon; 'Collier, Elaine (NIH/NCRR) [E]'Subject: RE: HIPAA Safe Harbor and implications on age over 90 in i2b2
Hi Shawn, Mike and thanks again to everybody interested, Yes, that's the standard approach and we do the same here now.
That said,
The 1000 genome project (actually the 1700) is holding highly personalized data on amazon AWS: http://aws.amazon.com/1000genomes/
a recent report from the Working Group on Data and Informatics to the NIH Advisory Committee to the Director mentions a couple places where cloud for images and Informatics/IT environments. http://acd.od.nih.gov/diwg.htm
There are also cloud based EMR vendors though I'd also be a bit leery of running a 24/7 clinical app on the cloud but it may be the best way to scale and offer value given the alternatives. For every cloud horror story there's a story like Cerner's DNS change severing access for many hours or unpublicized incidents at hospitals medical centers. When I was running clinical systems at Vandy we got better over time but always cringed at core network upgrades, DNS changes and other system changes taking us out. Here at KU it's been a virus software upgrade and a Citrix farm upgrade more so than network problems.
upgrades, DNS changes and other system changes taking us out. Here at KU it's been a virus software upgrade and a Citrix farm upgrade more so than network problems.
That's why i2b2 and other translational informatics seems pretty well suited to the cloud (amazon or rackspace).
The space is changing significantly over the last two years I've casually watched. Amazon also now has a group supporting university efforts in this area and Steve Elliot there says HIPAA BAA's are always at the top of the concerns.
Clinical research would seem to be a place you can try the new technology before you start deploying more mission critical apps.
We don't have much source transactional data beyond audit logs. I'd probably want to clone my audit logs back down to local storage.
If the whole thing gets destroyed, we rebuild or IDR every month anyway.
It's free to upload and you are charged on download ideal for us since we pull down only a tiny portion into data sets for analysis relative to building the whole instance.
Once we've got it up there, we can start doing heavy analysis and potentially exploit spot pricing for low cost
For bioinformatics/translational informatics, many of the public datasets may already be on the cloud, simplifying collaboration and reducing the need to copy data around between labs. If there weren't the regulatory concerns (and EMPI linkage challenges), it could be that clinical research informatics and public health could follow that lead with things like standard medicare/social security/tumor files available for linkage.
If you've got a DoD grant or need it to stay stateside, you can always pay the premium for govcloud to ensure it's managed in the US: http://aws.amazon.com/govcloud-us/
My sense is that Amazon runs and audits a data center and infrastructure well. Our institutional challenge is aligning our IT expertise to work with applications, scientists, and data sources to make sure we properly use that IT infrastructure whether it's in our basement or hosted elsewhere. If we can figure out how to do that, there's the potential to save money, speed deployment, and promote collaboration
Russ
From: Malin, Bradley A [b.malin.vanderbilt]
Sent: Monday, December 31, 2012 11:03 AM
To: i2b2 AUG Members; Nathan Graham; Russ Waitman
Subject: RE: HIPAA Safe Harbor and implications on age over 90 in i2b2
I, for one, will welcome Cloud-like solutions with open arms once the legal liability issues can be worked out... but since the initial question was not about the Cloud, I'll skirt this topic for now.
Russ, regarding your original question, this sounds tricky and I'm not sure I completely understand the situation. Perhaps this is because I'm not completely intimate with i2b2 (sorry Shawn!), but it may also be because there are multiple issues on the table. So, don't consider what I have to say here as the be-all-end-all of the discussion.
So, with that said, it sounds to me that you are only worried about the Safe Harbor and not the Expert Determination (or Statistical/Scientific standard) associated with the HIPAA de-identification model, so I will continue under this assumption. As Luke correctly pointed out, HIPAA de-identification does not require that the data be free and clear of risk, but if you know that the information could be identified - or fails to satisfy the de-identification requirements in a manner that is predictable and replicable - then it is certainly a violation of Safe Harbor in the eyes of OCR/HHS.
Regarding the specific examples that you offered, the 1918 influence case may or may not be deemed a violation. If:
i) 1918 was documented in clear and plain sight and designated as a date (as opposed to part of a proper noun) and
ii) there was additional documentation in clear and plain sight that the individual as alive at a future date that implied they were over 90 (e.g., a surgery in 2012) then again, yes, this would be a violation and you should either suppress the 1918 date to be "before [insert year here]" or change the 2012 date to be "after [insert year here]". I am not sure how i2b2 handles this issue however and would defer to Shawn.
But it sounds to me like the 1918 incident is rare and relatively unpredictable, in which case it is unlikely that this would be a de-identification violation. Actually, it's more likely that the fact the person had this variant of the flu would be more distinguishable than their age...
Anyway, going back to your options:
#1: Sounds fine, if you're willing to accept that the underlying DB is not de-identified. Of course, you have to be careful to ensure that you don't give out different versions of the same underlying data to the same investigators...
#2: I'm not a big fan of shifting some people and not others. I'd prefer generalization to say the individuals were born before year X. I really think the "1918" issue here is a special case in which knowledge is derived from the name of an event and not an explicit reporting of a date. It's certainly something that you want to be aware of and mitigate as such issues are known (e.g., redact 1918 from the name of the diagnosis), but I still think related situations may be considered "derived" and not "actual" knowledge under HIPAA.
#3: I really think this is where you need some business intelligence to handle the 90 year through generalization, not shifting.
Have to run, but it sounds like your problem could be solved by:
i) running a check on if the individual is known to be over 90.
ii) if they are over 90, then you either:
a) generalize all events before a certain date early in life (which might preclude the analysis of pediatric-related disorders)
b) generalize all events after a certain date later in life (which might preclude the analysis of geriatric-related disorders) However, this is why de-identified data is not necessarily conducive to all types of biomedical research...
brad
From: Darren W Henderson Sent: Monday, December 31, 2012 11:16 AM To: Mendis, Michael E. Cc: Luke Rasmussen; Russ Waitman; i2b2 AUG Members; b.malin.vanderbilt; Andrew Q Winter; Nathan Graham Subject: Re: HIPAA Safe Harbor and implications on age over 90 in i2b2
There is a white-paper on using Amazon Web services for HIPAA compliance available at http://aws.amazon.com/security/ down the page some.
Microsoft has also rolled out their features regarding HIPAA compliance in their azure cloud services for those that prefer MSSQL Server. http://www.windowsazure.com/en-us/support/trust-center/compliance/
I don't believe there has been time for legal precedent, but I would think there would be an insulating benefit for hosting PHI on a cloud service. If there was a security breach or leak on their secure cloud, the institution may avoid the full burden of the penalties levied for the loss of PHI. I know at our institution we spend a great deal of time and resources to try and prevent such a thing from happening, but we are hardly ready to handle those penalties if there were to be a problem.
From: Murphy, Shawn N. [SNMURPHY.partners]
Sent: Monday, December 31, 2012 10:19 AM
To: Mendis, Michael E.; Russ Waitman
Cc: Luke Rasmussen; Malin, Bradley A; i2b2 AUG Members; Nathan Graham; Andrew Q Winter
Subject: RE: HIPAA Safe Harbor and implications on age over 90 in i2b2
Mike implied this, but just to be clear, we don't put any real patient data, de-identified or not, on the cloud right now from Partners Healthcare. Data replication practices in the cloud are certainly one important reason, and ambiguity of encryption key guarding practices is another.
I do see a solution coming within the next two or three years, and would certainly expect this community to be forging the way.
Thanks,
Shawn.
From: Mendis, Michael E.
Sent: Monday, December 31, 2012 10:54 AM
To: Russ Waitman
Cc: Luke Rasmussen; b.malin.vanderbilt; i2b2 AUG Members; Nathan Graham; Andrew Q Winter
Subject: Re: HIPAA Safe Harbor and implications on age over 90 in i2b2
Russ,
On a side about the amazon cloud. We use it for the community site, the actual i2b2 demo and a few side i2b2 projects. For these it has been very useful and beneficial with working with other groups.
A few pitfalls that we ran into, a few months ago during a storm in Virginia the oracle database crashed, Lost power. They were unable to restore the DB and as a result we had to rebuild the demo data, not a big deal for us, this was the major RDS from amazon, not the express edition included on the vms. Another thing, even those you say use the data storage on the east coast, you can't guarantee that the data might be on a backup somewhere in china. iirc, with clinical trials and FDA the data has to stay within the US. We dont use any encryption on the data, but even if we did, we also can't guarantee that core dumps and such might be saved somewhere which could contain unencrypted data.
Mainly what I am getting at, with amazon and all these public clouds, is there is no guarantee that the data might escape out.
mike
From: Malin, Bradley A [b.malin.vanderbilt] Sent: Monday, December 31, 2012 12:06 PM To: Russ Waitman Cc: Nathan Graham; i2b2 AUG Members Subject: RE: HIPAA Safe Harbor and implications on age over 90 in i2b2
Right-o. Age over 89 and any dates that may imply such information is explicitly designated under §164.514(b)(2) (C).
(C).
brad
From: Russ Waitman [rwaitman.kumc] Sent: Monday, December 31, 2012 9:30 AM To: Matvey Palchuk; Luke Rasmussen; Malin, Bradley A; i2b2 AUG Members Cc: Nathan Graham; Andrew Q Winter Subject: RE: HIPAA Safe Harbor and implications on age over 90 in i2b2
Hi Matvey,As a point of clarification, I think it is required by HIPAA. See page 766. 164.514.b.2.i.Chttp://www.gpo.gov/fdsys/pkg/CFR-2007-title45-vol1/pdf/CFR-2007-title45-vol1-chapA-subchapC.pdf Russ
From: Matvey Palchuk [MPalchuk.recomdata] Sent: Monday, December 31, 2012 8:48 AM To: Russ Waitman; Luke Rasmussen; b.malin.vanderbilt; i2b2 AUG Members Cc: Nathan Graham; Andrew Q Winter Subject: Re: HIPAA Safe Harbor and implications on age over 90 in i2b2
I'll add my $0.02...
if you set out to build de-identified data set (per HIPAA), all 18 PHI elements have to be excluded. These 18 include dates and ZIP codes. Dates (all dates – DOB, DOD, start and end dates, etc.) must be stripped down to a year (as in, everything happens on 1st of January, for example). There has been a precedent of IRBs accepting a shift (either adding or subtracting) by a random number from 1 to 365 (I personally would exclude 0 from this interval) instead. ZIP codes must be stripped down to 3 digits and of a geographic area described by these 3 digits contains a population <20K, that ZIP must be reported as 000.
As you know, in addition to excluding 18 PHI elements, HIPAA has language about covered entity's knowledge that the identity of the person may still be determined based on whoever data is provided – if such knowledge exists, the data set can not be considered de-identified.
Age > 90 – this is not a HIPAA provision; it's from NIH Common Rule. It is our practice to use a rule like "age>89 = 90" when loading i2b2
Regarding access to patient dimension – this brings up an interesting nuance. So far we talked about building the data mart and being within HIPAA safe harbor. Now let's talk about user access. Assuming you do not let your users access the database directly, they are limited by whatever role you provide in i2b2. At "obfuscated" level of access, users can not save a patient set, can not use plug-ins, and so regardless of whether the underlying data mart has de-identified or limited data set, the user has "functional" access to de-identified data only by virtue of restrictions imposed by "obfuscated" role.
Matvey
Matvey B. Palchuk, MD, MS
CMIO, Recombinant By Deloitte.
+1 617 243 3700 x218 | Mobile +1 781 790 1590 | Fax +1 855 318 1185
On Dec 31, 2012, at 9:29 AM, Russ Waitman wrote:
Hi Luke,
We're doing #1 basically but I am concerned if I reveal facts separated by more than 90 years, I wonder if that's a concern of HIPAA. I still am not revealing an explicit age though.
The business issue I have in the short term is that I'd like to begin to be able to host solutions at places like Amazon.com which won't sign a HIPAA Business Associates Agreement. But if my data is fully de-identified and determined non-human subjects (and we use Oracle encryption), I and the contributing organizations don't need to require that. In the long run, I think this approach may save us significant money and help us support other organizations.
Thanks for the link to the slides. I enjoyed that session.
Russ
From: Luke Rasmussen
Sent: Saturday, December 29, 2012 10:34 PM
To: Russ Waitman; b.malin.vanderbilt<b.malin.vanderbilt>; i2b2 AUG Members<i2b2 AUG Members>
Cc: Nathan Graham; Andrew Q Winter
Subject: Re: HIPAA Safe Harbor and implications on age over 90 in i2b2
Russ and Nathan -
I'm far from an expert in this area, and am very interested to hear what others (especially Brad) have to say on the matter. Just my $0.02 given what you've stated. If you are trying to keep a HIPAA de-identified data set, your solution #1 seems the most appropriate, since you note the patient dimension table is technically not de-identified. Is there a strong business need to grant access to that table? Depending on what questions people are asking of the data, you could always synthesize new facts to answer them - are they wondering how old someone was when they died? Is it just simply the mortality status? Is it death in relation temporally to other clinical events?
As I'm sure Brad and others can speak to in greater detail, you can follow the HIPAA de-identification rules and still potentially be at risk for re-identification. I think it's good you're taking the time to explore the different angles. And to answer your question - I do lose some sleep worrying about honest broker issues, and hope I'm not the only one! It's really tough when you have people making a strong case to get more specific, granular data, but it potentially conflicts with appropriately securing the data.
The slides from an AMIA panel on honest brokers & de-identification may also be of interest: https://vault.it.northwestern.edu/lvr491/public_html/S69-AMIA2012-Final.pdf
Looking forward to hearing what others have to say on the matter.
Luke Rasmussen
Systems Analyst/Programmer
Northwestern University
Division of Health and Biomedical Informatics
(312) 503-2823
From: Russ Waitman
Date: Friday, December 28, 2012 4:53 PM
To: b.malin.vanderbilt, i2b2 AUG Members
Cc: Nathan Graham
Subject: HIPAA Safe Harbor and implications on age over 90 in i2b2
Dear i2b2 and Brad (the health privacy guru),
We've run into an interesting scenario as we accumulate data in our i2b2 based integrated data repository which has implications for how we manage access and release data in a fully de-identified manner.
Background:
We've attempted to create fully deidentified data repository by stripping out all 18 identifiers her HIPAA Safe Harbor and we then randomly shift birthdays and all dates for each patient by the same 0 to negative 365 day offset. This is helpful with building consensus across the contributing organizations and may also set us up nicely to manage the data as non-human subjects data when it comes to technical infrastructure.
By HIPAA safe harbor, we have to manage people over 90 in a special category. Aka, we can't tell you they are 92 or 104.
When load i2b2 facts, our ETL process caps all age facts at 90 years old.
Situation:
We realize now though that's we've populating the patient dimension with the real birthday and we added death via social security or the hospital records as a fact. So, if we give access to the patient dimension and the "death fact", people can extrapolate patients real age as both birthdate and death date are offset by the same amount, thus revealing people over 90.
Solutions?
1. Never give out the patient dimension..... but in a way, our de-identified repository isn't really fully compliant with the over 90 clause due to the fact that we contain the birthdate on people over 90. But we can enforce we only give out facts not the patient dimension so we could enforce this technically to prevent disclosure.
What do people think about that from a HIPAA point of view?
2. Shift the birth date forward so every one is 90 years old or younger. Of course if you died a 89, no need shift you even if you died in 1965.
Problems with approach #2
If we don't have a death record, there's the potential for significant shifting which will also change as we refresh the data.
Bad case example: Someone is born in 1910, had a record that they were afflicted by the 1918 influenza, they may still be alive ... or dead... we don't know.
If we shift their birthdate up to 1923 to make them less than 90, then the influenza fact from 1918 either reveals their age because it's a fact before they are born, or we have to also shift facts more than 90 years ago which gets messy and possibly computationally intensive as it goes against the fact table. Never to mind the fact that we'd lose record of things like the 1918 influenza as an absolute public health event.
3. Shift birthdays and facts datetimes forward in time so that there's no interval of time for a patient in the database greater than 90 years.
Even worse case: it's theoretically possible in the example above that we could have two facts about a person more than 90 years apart. Their past medical history denotes the 1918 influenza and then here they are in 2012 undergoing a hip fracture procedure. We don't even have to disclose their age to know that they are more than 90 years old. (We'll check).
Are we over interpreting HIPAA? Any one else stay awake at night worried about honest broker concerns?
Thank you for your thoughts and we hope Santa brought everyone the new hardware for their data repositories they wanted for Christmas,
Russ Waitman and Nathan Graham
University of Kansas Medical Center
RxNorm ontology [12-28-2012]
From: Russ Waitman [rwaitman.kumc] Sent: Friday, December 28, 2012 3:43 PM To: Eunjung S Lee; i2b2 AUG Members Cc: Nathan Graham Subject: RE: RxNorm ontology
Hi Sally,We use NDF-RT and spent a fair amount of time deciding how to bundle meds together in i2b2 and read an article by Matvey Palchuk Enabling Hierarchical View of RxNorm with NDF-RT Drug Classes. Palchuk MB, Klumpenaar M, Jatkar T, Zottola RJ, Adams WG, Abend AH NDF-RT is the proposed source of drug classification information. We set out to construct a hierarchy of NDF-RT drug classes and RxNorm medications and evaluate it on medication records data. NDF-RT and RxNorm are distributed in different file formats, require different tools to manipulate and linking the two into a hierarchy is a non-trivial exercise. Medication data in RxNorm from two institutions was constrained by the hierarchy. Only 37% of records from one and 65% from another institution were accessible. We subsequently enriched the RxNorm mapping in NDF-RT by exploiting relationships between concepts for branded and generic drugs. Coverage improved dramatically to 93% for both institutions. To improve usability of the resulting hierarchy, we grouped clinical drugs by corresponding clinical drug form. PMID: 21347044 PMCID: PMC3041416URL - http://www.ncbi.nlm.nih.gov/pubmed/21347044?dopt=Citation Our original med data from our Epic EMR is tagged with FDB or NDC codes. Nathan Graham led most of the work and spent considerable time trying to get coverage of medications which didn't map to RxNorm. Since we did our reinterpretation as part of our CTSA, our work is all freely available on our website Some notes are herehttps://informatics.kumc.edu/work/wiki/MedMapping Much of Nathan's work is in these ticketshttps://informatics.kumc.edu/work/ticket/1048https://informatics.kumc.edu/work/ticket/1246 and the code is in our main heron ETL folderhttps://informatics.kumc.edu/work/browser/heron_load specifically https://informatics.kumc.edu/work/browser/heron_load/epic_med_mapping.sqlmaps the meds to the NDF-RT ontology whilehttps://informatics.kumc.edu/work/browser/heron_load/epic_meds_transform.sqlloads the facts and extra modifier features like the estimated daily dose. You'll want to download NDF-RT and RxNorm from the UMLS. If you have questions, feel free to email Nathan and I.
Russ Waitman, PhD
Director of Medical Informatics
Assistant Vice Chancellor for Enterprise Analytics
Associate Professor, Department of Biostatistics
913-945-7087 (office)
rwaitman.kumchttp://informatics.kumc.edu
From: Mendis, Michael E. Sent: Friday, December 28, 2012 3:32 PMTo: Lee, Eunjung S; i2b2 AUG MembersSubject: RE: RxNorm ontology
Emily,
The rxNorm that you see on the portal is in a single level ontology, which is why you cant see it on the tree, there are about 200K of entries. What I think that you want to do is take the NDC code and find the Drug Products by VA Class level. That will contain a NUI code, and than you will want to cross walk between the NDC and rxnorm codes.
But yeah extract the NDC from the bioportal and use the RxNorm flat file from the nih
mike
From: Eunjung S Lee [sallylee.u.washington] Sent: Friday, December 28, 2012 1:57 PM To: Russ Waitman; i2b2 AUG Members Subject: RxNorm ontology
Hello,
I am wondering if anyone might have a good rxNorm medication ontology or know where to find one.
I have one that is rxnorm based (Drug Products by VA Class), but that did not match many of the valid rxcui codes in my dataset so I think I will need a more detailed one.
I went to find one in bioportal and found one promising (http://bioportal.bioontology.org/ontologies/49463?p=terms), but it won't display the tree so I don't know if I would be able to extract it using the extraction tool (although I have not used this before so maybe there is a trick to it).
Any help would be great since I'm still new to this.
Thank you!
Sally
E. Sally Lee, Ph.D
Biomedical Informatics Core Consult
Amalga Clinical Data Repository
Institute of Translational Health Sciences
University of Washington
sallylee.uw
Age calculation [12-21-2012]
From: Wanta Keith M [KWanta.uwhealth] Sent: Friday, December 21, 2012 3:43 PM To: 'Wilson Lau'; i2b2 AUG Members Subject: RE: Age calculation
You are correct Wilson.
I'm guessing not too much thought went into this algorithm because an organization will generally first use i2b2 as a cohort tool (for getting patient counts) using a de-identified patient data set. In a de-identified patient data set, all 18 forms of PHI must be removed from your fact values (based on your ontology concepts) within the i2b2 OBSERVATION_FACT table, when following HIPAA. In the link below, you will notice that #3 describes not to expose dates associated to the patient. Year is okay. Generally, organizations will have the age (year) of the patient in their demographics ontology. Even then, it's not extremely precise. It's also good practice to shift any dates (ie: service dates---date of fact, birth date) randomly by some number of days to hide the details of the fact in order to protect patient privacy. You must also remove any patients older than 89.
http://cphs.berkeley.edu/hipaa/hipaa18.html
If you are populating a limited data set within i2b2, and want to expose precise dates, then it might be worth spending time on this algorithm. For example, maybe your organization wants to use i2b2 for pediatrics. In that case, the age of the patient needs to be more precise (1 month, 3 months, etc..) , then it gets more tricky so you can get more precise counts.
Keith Wanta • Data Warehouse Specialist, Associate
UW Health • HIMC • 608.890.5778
From: Wilson Lau [wlau.uw.edu] Sent: Tuesday, December 18, 2012 4:46 PM To: i2b2 AUG Members Subject: Age calculation
Hello,
I am using the demographics ontology from the demo data.
The generated SQL for Age calculation is using 365.25 days per year.
For example,
SELECT /*+ index(observation_fact fact_cnpt_pat_enct_idx) */ patient_num
FROM dbo.patient_dimension
WHERE patient_num IN (
SELECT patient_num
FROM dbo.patient_dimension
WHERE birth_date BETWEEN getdate() - (365.25 * 2) + 1
AND getdate() - 365.25 + 1
)
When I look at the range of the dates in the where clause, I have the following data
SELECT GETDATE() AS [Now]
, 1 AS [Age]
, GETDATE() - (365.25 * 2) + 1 as [begin]
, GETDATE() - 365.25 + 1 as [end]
Now |
Age |
begin |
end |
2012-12-18 14:32:30.773 |
1 |
2010-12-20 02:32:30.773 |
2011-12-20 08:32:30.773 |
If a patient was born 2011-12-19, shouldn't he/she be considered age of 0 ?? But based on the i2b2 calculation, that would be age 1.
Thanks.
Wilson
May be useful to someone out there [12-20-2012]
From: Dan Connolly [dconnolly.kumc] Sent: Thursday, December 20, 2012 3:32 PM To: Russ Waitman; 'Jack London'; 'Luke Rasmussen'; i2b2 AUG Members Subject: RE: May be useful to someone out there.
I did a blog item on it way back when:
Concept stats: how many needles are in which parts of our i2b2 haystack?
Posted: 2010-12-16 It's somewhat tricky to maintain, but we find it worth the trouble. Better integration with the c_totalnum stuff in recent i2b2 releases is #779 on our todo list. It's been there for a year. Every monthly release I seem to run out of round 'tuits before I get it done. Sigh.
From: Russ Waitman [rwaitman.kumc] Sent: Thursday, December 20, 2012 2:01 PM To: 'Key, Dustin'; 'Harle,Christopher A'; 'Luke Rasmussen'; i2b2 AUG Members Subject: RE: May be useful to someone out there.
Yes. We show the full ontological path in the tool tip so when people search terms they can also go back and find the terms position in the ontology
It works well most of the time
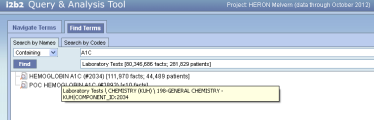
But I think we have issues when the path length relative to the tool tip field size when the paths are real verbose ala CPT (see below).
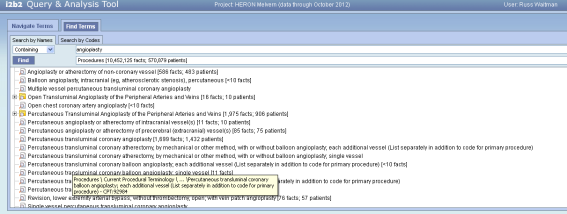
Russ
From: Key, Dustin [key.d.ghc] Sent: Thursday, December 20, 2012 11:18 AM To: 'Harle,Christopher A'; Russ Waitman; 'Luke Rasmussen'; i2b2 AUG Members Subject: RE: May be useful to someone out there.
We took the subcategory approach with different kinds of procedure types, separately. And Chris, I too like including the codes. In cases such as Rx, without a numeric sort order, users have had difficulty finding terms in the hierarchy tree, after they have found them in the search terms. (One idea I've had is to perhaps put the first few branches of each node in the tool tip field so that when a user finds a term via search terms, they can navigate back to it by starting at the tree root. Have others tried this? ) Also, I really like what Russ's team has done with the people/fact count built into the ontology. Dustin

From: Harle,Christopher A [charle.phhp.ufl] Sent: Thursday, December 20, 2012 8:51 AM To: 'Russ Waitman'; 'Luke Rasmussen'; i2b2 AUG Members Subject: RE: May be useful to someone out there.
Russ,
Thanks, this is helpful to see what others are doing and your thought processes. In our i2b2 ontology, I've insisted on including actual codes (icd9, cpt, etc.) directly in the visible names. My rationale was the codes were needed to help users (especially non-clinicians like myself) get their bearings. I was concerned that too much use of the find terms function would lead people not to go back to the hierarchy and end up missing relevant terms. But, like you, I am without empirical evidence on what is actually intuitive. We could evaluate that of course!
Chris
From: Russ Waitman [rwaitman.kumc] Sent: Thursday, December 20, 2012 11:31 AM To: 'Luke Rasmussen'; i2b2 AUG Members Subject: RE: May be useful to someone out there.
Well I am not sure it's necessarily worked well but it was the easiest thing to do. Until we get a bunch of usage out of i2b2 by a cross section of different kinds of researchers, it's not clear to me we really understand how intuitive the various ontologies really are for people.
I find myself always using i2b2 side by side with google and http://icd9cm.chrisendres.com/ to search for codes, find the code's position in the ontology by find terms on code, and then seeing what other related concepts are there.
Russ
From: Luke Rasmussen [luke.rasmussen.northwestern] Sent: Thursday, December 20, 2012 10:25 AM To: Russ Waitman; i2b2 AUG Members Subject: Re: May be useful to someone out there.
Thanks Russ! It's good to hear how this approach worked – it was one we had discussed, but weren't sure how people would react.
From: Russ Waitman [rwaitman.kumc] Sent: Thursday, December 20, 2012 12:25 PM To: 'Jack London'; 'Luke Rasmussen'; i2b2 AUG Members Cc: Dan Connolly Subject: RE: May be useful to someone out there.
It's "stored" in the text string of the ontology. Dan Connolly did it on our side when we were on 1.4 before we knew about i2b2 adding a slot for it.
I think the code is here: https://informatics.kumc.edu/work/browser/heron_load/concept_stats.sql
Russ
From: Jack London [Jack.London.jefferson] Sent: Thursday, December 20, 2012 10:49 AM To: Russ Waitman; 'Luke Rasmussen'; i2b2 AUG Members Subject: RE: May be useful to someone out there.
Russ,
How do you get the BOTH the number of facts and number of patients to appear in the metadata ontology display? We run a program after every refresh to set the c_totalnumber column in the metadata tables, but this only reports number of patients. Where do you store the number of observations for display?
Jack
From: Russ Waitman <rwaitman.kumc> Date: Thursday, December 20, 2012 9:40 AM To: Luke Rasmussen <luke.rasmussen.northwestern>, i2b2 AUG Members Subject: RE: May be useful to someone out there.
At this point we have them both under "procedures" but they are in separate sub-ontologies. May not be ideal but haven't had much feedback from people. For the couple customers we have had in this area they tend search by the CPTs they know for some prospective trial cohort identification.
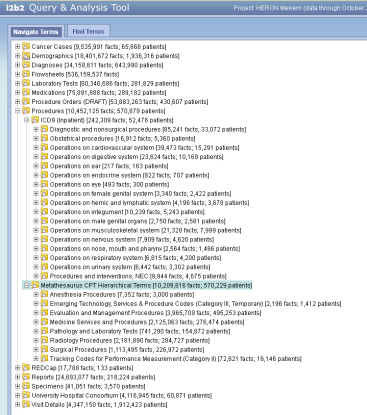
Russ
From: Luke Rasmussen [luke.rasmussen.northwestern] Sent: Wednesday, December 19, 2012 4:49 PM To: Russ Waitman; i2b2 AUG Members Subject: Re: May be useful to someone out there.
What Darren had provided to pull procedure and diagnosis codes from the UMLS was a great starting point for us. I was wondering if anyone has run into a scenario where you have both ICD9 and CPT codes for procedures, and want to place them in the same procedure hierarchy in i2b2. Essentially I would like (if possible) to allow a user to navigate to drag over a single "Prostate surgery" folder in the ontology and have it query both ICD9 Px and CPT terms.
I appreciate any feedback if anyone has tried this, thinks this is a terrible idea, or may have already solved this issue.
Thank you,
Luke Rasmussen
Systems Analyst/Programmer
Northwestern University
Division of Health and Biomedical Informatics
(312) 503-2823
From: "Henderson, Darren W" <darren.henderson.uky> Date: Wednesday, February 15, 2012 9:31 AM To: i2b2 AUG Members Subject: May be useful to someone out there.
We recently needed to update our ICD9 and CPT ontologies in i2b2 when we found some missing codes that were fairly new. We were missing codes for MRSA and some other random things. So I tossed this together to pull the i2b2 ontology out of the UMLS tables mrhier and mrconso that you can download out of the umls and then import into a database to work with. I figured I'd toss this out into the members list wind so no one has to waste time reinventing the wheel if you happen to need an update to your ontology. Cheers.
Darren W. Henderson
Institute for Pharmaceutical Outcomes and Policy
Center for Clinical and Translational Science
Division for Biomedical Informatics
University of Kentucky
789 S. Limestone Rm. 182
Lexington, KY 40536
(859) 323-7146
(859) 967-4914
Concept path containing '<' [12-17-2012]
From: Mendis, Michael E. Sent: Monday, December 17, 2012 3:19 PMTo: Yves ThorrezCc: i2b2 AUG MembersSubject: Re: Concept path containing '<'
We are trying to reproduce this on our end.
thanks
mike
On Dec 17, 2012, at 11:07 AM, Yves Thorrez wrote:
Hello,
An error occurs upon running any query where the concept path contains a '<' (checked for '>' and that's ok though). We're running the i2b2 v1.6.07. Included is a partial server log for an example.
Thanks for any feedback,
Yves
Access control in i2b2 hierarchy [12-11-2012]
From: Liu,Felix [felix.liu.ufl] Sent: Tuesday, December 11, 2012 10:04 AM To: Michael Horvath; 'i2b2 AUG Members' Subject: Re: Access control in i2b2 hierarchy
I have toyed with this idea and presented a poster describing a clumsy way at AMIA 2012:
Deason R, Lipori G, Ebel C, Liu J. Research Specific Data Management using an I2B2 Data Warehouse. AMIA Annu Symp Proc. 2012 Nov 3:1716.
If your backend database support it (e.g., Oracle), you can probably try to use Virtual Private Database to achieve the same thing, provided I2B2 query tool pass user credentials to the DB. I believe VPB will cost quite a bit of $$. I haven't tried VPB myself.
Felix
J. Felix Liu, Ph.D.
Director, CTS-IT Program
Clinical and Translational Science Institute
University of Florida
Email: felix.liu.ufl
Phone: (352) 273-8119
From: Murphy, Shawn N. Sent: Tuesday, December 11, 2012 9:38 AM To: Michael Horvath; 'i2b2 AUG Members' Subject: RE: Access control in i2b2 hierarchy
Hi All,
The prescribed way to limit the ability of a group of users to a specific set of ontology items is to create a project for them, and then associate them with the specific ontology database through the project path. Many projects can then point to the same underlying data repository but only allow querying of what is in the ontology cell for each specific project. The server will check with the project ontology cell to make sure that ontology item that is used in a query is indeed allowed by that project. That way, a person cannot "hack" the system and put forbidden items in the xml directly, and everything is consistent (query in query, etc.)
Thanks,
Shawn.
From: Michael Horvath <mhorvath.wakehealth> Date: Monday, December 10, 2012 12:49 PM To: "'i2b2 AUG Members'" <i2b2 AUG Members> Subject: RE: Access control in i2b2 hierarchy
I've not done it, but shouldn't you be able to associate a cell for a project with datasources that contain a subset of the data (you may have to specify multiple datasources for the all inclusive cell)? This way shouldn't require that you duplicate any data. You can then use project_path's to control which cells get picked up for a project (the hive will return the cell that is most specific to the project path).
From: Russ Waitman [rwaitman.kumc] Sent: Monday, December 10, 2012 12:31 PM To: 'Peter Beninato'; 'i2b2 AUG Members'; Bhargav Adagarla Subject: RE: Access control in i2b2 hierarchy
Hi Peter,
We don't want to have to clone all the EMR data in each project. We want to blend the small registries together with the large EMR and billing records (say 250GB to 1TB),
Russ
From: Peter Beninato [beninato.ohsu] Sent: Monday, December 10, 2012 11:11 AM To: Russ Waitman; 'i2b2 AUG Members'; Bhargav Adagarla Subject: RE: Access control in i2b2 hierarchy
Hi,
Perhaps separate Projects need to be established in your i2b2. This would allow grants be done on those projects to whoever needed access, and prevent others from not seeing it.
From: Bhargav Adagarla [badagarla.kumc] Sent: Monday, December 10, 2012 8:31 AM To: 'i2b2 AUG Members' Subject: Access control in i2b2 hierarchy
Hello all,
Earlier this year we started bringing data from projects in REDCap, into i2b2 (https://informatics.kumc.edu/work/browser/heron_load/redcap_i2b2_transform.sql). This work made us realize the need for finer access control in i2b2 : controlling which folders the users can access when they log onto i2b2. For e.g. (in the picture below), hiding the folder Circulatory System under Diagnoses from the logged in user depending on whether he has rights to the data under the folder.
This would be useful in limiting access to i2b2 data (from projects in REDCap) to certain users.
We were wondering if any of you have implemented or have tried to implement finer access control in i2b2, like this. Any thoughts and suggestions will be greatly appreciated. Thanks.
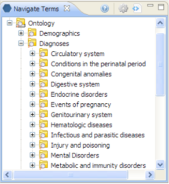
(Source: i2b2 documentation)
Regards,
Adagarla, Bhargav Srinivas
University of Kansas - Medical Center
913-588-0447
UMass Med School i2b2 Web Client plug-ins & DBA Toolset for the FAIR Initiatve [12-03-2012]
From: Chan, Wayne [Wayne.Chan.umassmed] Sent: Monday, December 03, 2012 10:09 AM To: 'i2b2 AUG Members' Cc: Sadasivam, Rajani; Houston, Thomas; English, Thomas Subject: The UMass Med School i2b2 webclient plugins & DBA Toolset for the FAIR Initiative
Hi, all,
We at the University of Massachusetts Medical School are happy to share with the rest of the i2b2 community the FAIR (Familial, Associational, and Incidental Relationships) i2b2 webclient plugins and DBA Toolset that were announced in the recent CTSA iKFC Meeting. They are available for download in our corresponding website page, http://micard.umassmed.edu/software.html (near the bottom).
We have also set up corresponding i2b2 Community project Wiki pages (https://community.i2b2.org/wiki/display/FAIR/Home) about these new tools. This is the link at the dashboard page:

Please don't hesitate to contact us concerning any questions or problems. Thanks.
Wayne
S. Wayne Chan, MSEE, MSME
Biomedical Research Informatics Development Group (BRIDG) and Biomedical Research Informatics Consultation & Knowledge Service (BRICKS),
Division of Health Informatics & Implementation Science (HIIS),
Department of Quantitaive Health Sciences (QHS),
University of Massachusetts Medical School (UMMS) at Worcester, MA 01655
(508) 856-8947





