- Dashboard
- Ontology Tools
- Ontology Tools
- NCBO Extraction tool version 1.0
- Attachments
- i2b2 Ontology Tutorial.doc
i2b2 Ontology Tutorial.doc
i2b2 Ontology Tutorial
Follow along for a step by step example of how metadata trees are created and configured.
i2b2 Navigate Terms View
Creating the root level nodes
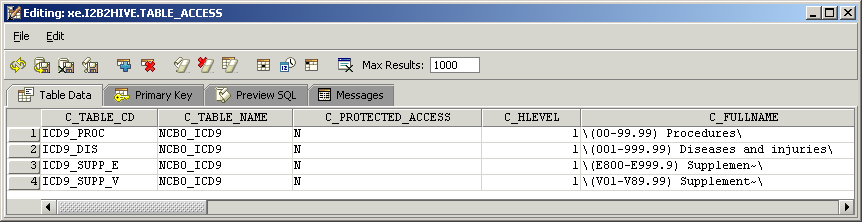
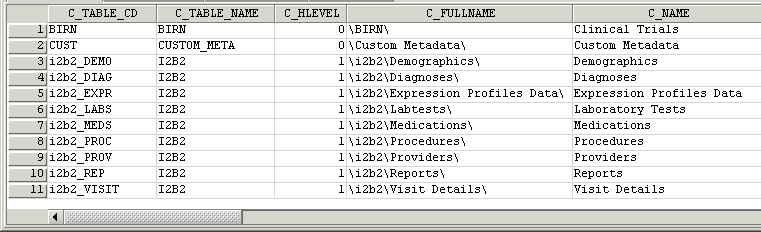
There is a one-to-one mapping between the entries of the table_access table and the root level nodes for a given project.
Building local metadata
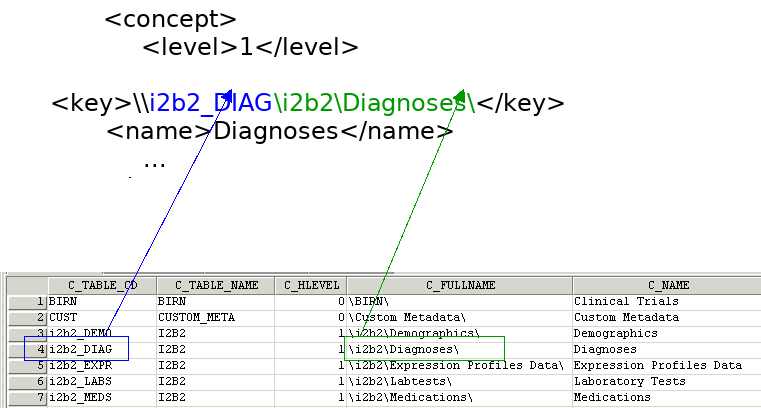
A concept’s key is made up of two parts: \\c_table_cd\c_fullname
The c_table_cd tells the ONT cell which metadata table the concept resides in (‘i2b2’), while c_fullname is a unique identifier for the concept itself.
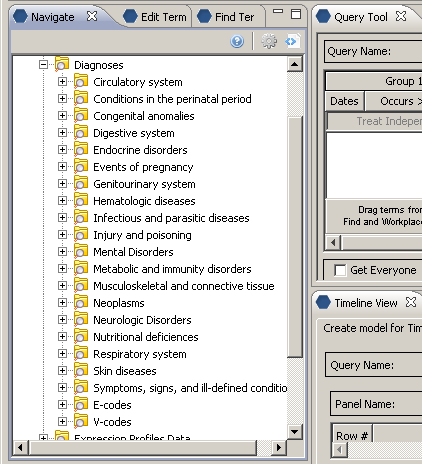
Children of concept Diagnoses [\i2b2\Diagnoses\]
C_HLEVEL C_FULLNAME C_NAME
----------- ---------------------------------------------------------------------------------------- -----------------------------------------------------------
2 \i2b2\Diagnoses\Circulatory system (390-459)\ Circulatory system
2 \i2b2\Diagnoses\Conditions in the perinatal period (760-779)\ Conditions in the perinatal period
2 \i2b2\Diagnoses\Congenital anomalies (740-759)\ Congenital anomalies
2 \i2b2\Diagnoses\Digestive system (520-579)\ Digestive system
2 \i2b2\Diagnoses\Endocrine disorders (240-259)\ Endocrine disorders
2 \i2b2\Diagnoses\Events of pregnancy (630-677)\ Events of pregnancy
2 \i2b2\Diagnoses\Genitourinary system (580-629)\ Genitourinary system
2 \i2b2\Diagnoses\Hematologic diseases (280-289)\ Hematologic diseases
2 \i2b2\Diagnoses\Infectious and parasitic diseases (001-139)\ Infectious and parasitic diseases
2 \i2b2\Diagnoses\Injury and poisoning (800-999)\ Injury and poisoning
2 \i2b2\Diagnoses\Mental Disorders (290-319)\ Mental Disorders
2 \i2b2\Diagnoses\Metabolic and immunity disorders (270-279)\ Metabolic and immunity disorders
2 \i2b2\Diagnoses\Musculoskeletal and connective tissue (710-739)\ Musculoskeletal and connective tissue
2 \i2b2\Diagnoses\Neoplasms (140-239)\ Neoplasms
2 \i2b2\Diagnoses\Neurologic Disorders (320-389)\ Neurologic Disorders
2 \i2b2\Diagnoses\Nutritional deficiences (260-269)\ Nutritional deficiences
2 \i2b2\Diagnoses\Respiratory system (460-519)\ Respiratory system
2 \i2b2\Diagnoses\Skin diseases (680-709)\ Skin diseases
2 \i2b2\Diagnoses\Symptoms, signs, and ill-defined conditions (780-799)\ Symptoms, signs, and ill-defined conditions
2 \i2b2\Diagnoses\zz E-codes\ E-codes
2 \i2b2\Diagnoses\zz V-codes\ V-codes
Concept Diagnoses’ children all start with a c_fullname of ‘\i2b2\Diagnoses\’
and all reside in the same table as ‘Diagnoses’ (c_table_cd = ‘i2b2_DIAG’, c_table_name = ‘i2b2’)
Children of concept Diagnoses display
Creating metadata for standard ontologies using BioPortal
NCBO BioPortal hosts over 250 ontologies
Any of these ontologies may be extracted for use within i2b2 through use of a standalone Extraction tool.
Browse ontologies within BioPortal and locate ontology of interest: (double click on name of ontology)
![]()
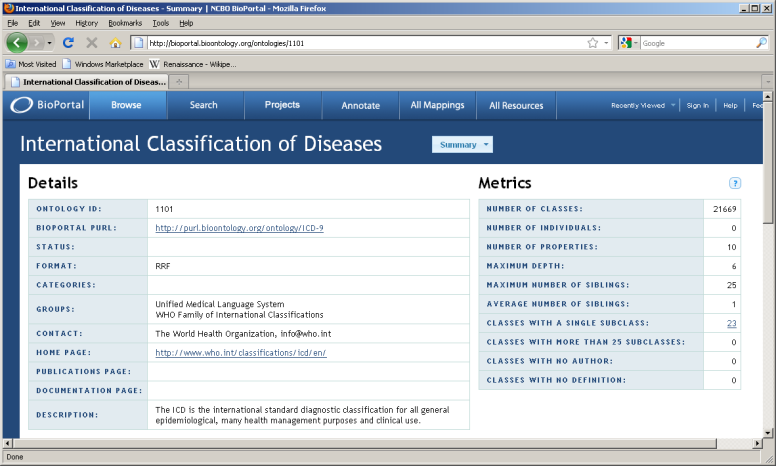
Detail summary page:
Locate ontology id for version of interest [45221]

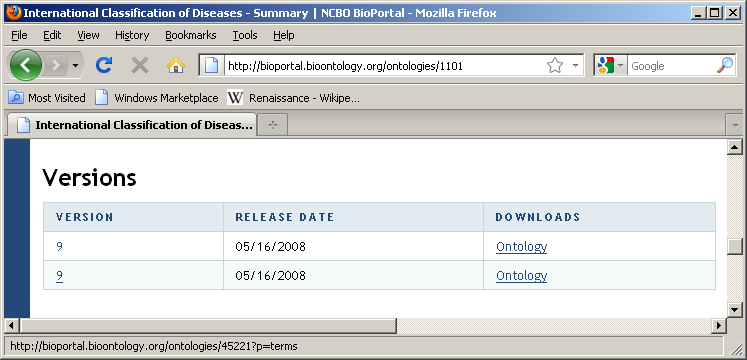

45221 is the ontology id which uniquely identifies this version of this ontology. It is generally a 5-digit number.
NCBO-specific Ontology Extraction tool inputs:
Ontology id: 5-digit id assigned to ontology version of interest
apikey: An API key assigned to you by NCBO. Log onto BioPortal (or get an account); your apikey is located on your Account page.
NCBO Ontology Extraction Workflow (2-stage process)
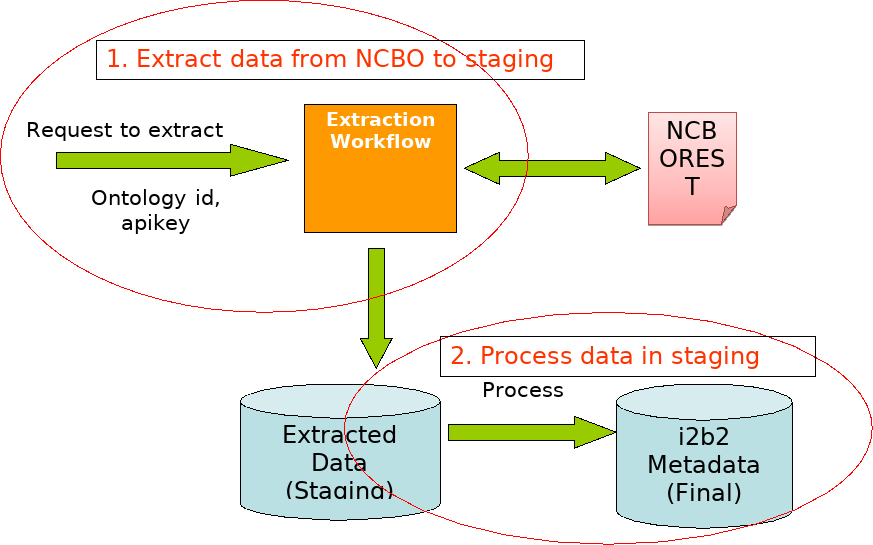
Point table_access to root nodes of your new extracted metadata.
Creating project-specific (localized) metadata
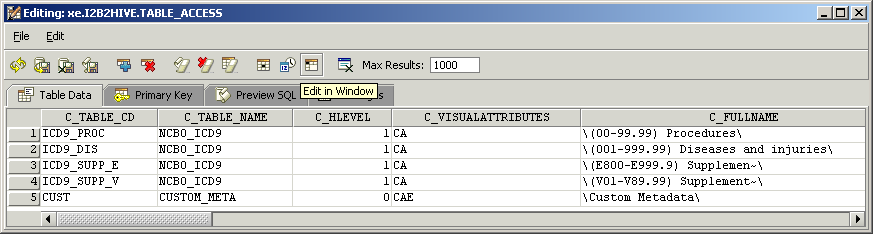
In some cases you may need to create a small, non-standard ontology for a local project. Consider the case for classifying patients as smokers, non-smokers or smoking status unknown. To support this effort, we create a new table ‘Custom_Meta’ whose root level entry ‘Custom Metadata’ has visual attribute ‘CAE’. The ‘E’ stands for ‘editable’ and makes the entry available for editing within the Edit Terms tool.
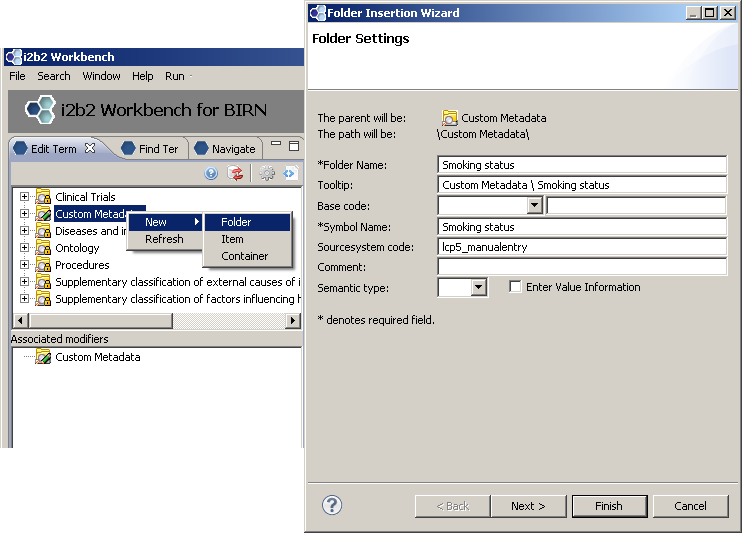
First we create a new folder “Smoking status”
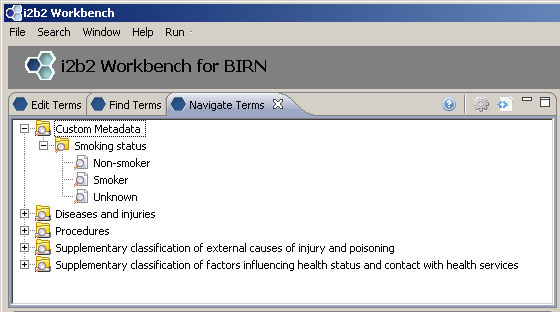
Add three concepts: “Smoker”, “Non-smoker”, “Unknown”
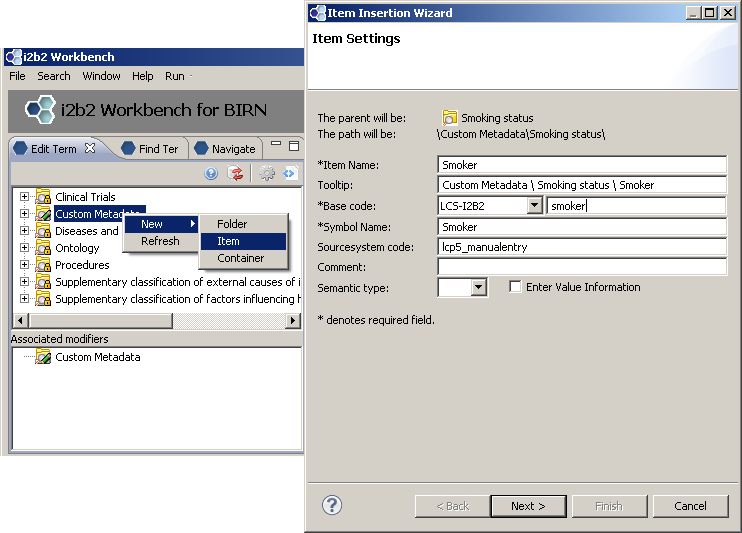
New folder with three new concepts:
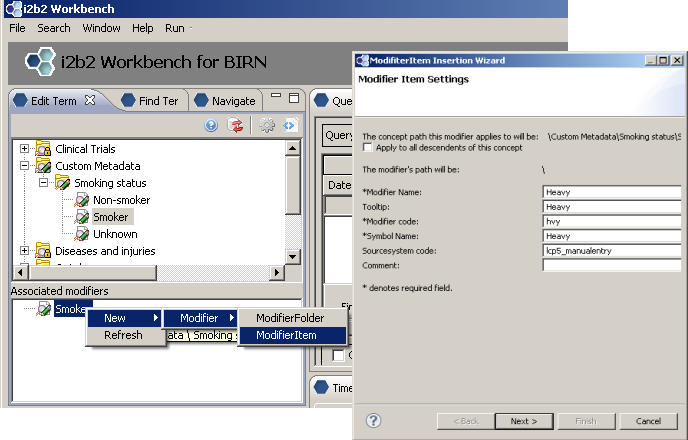
Creating modifiers for existing metadata
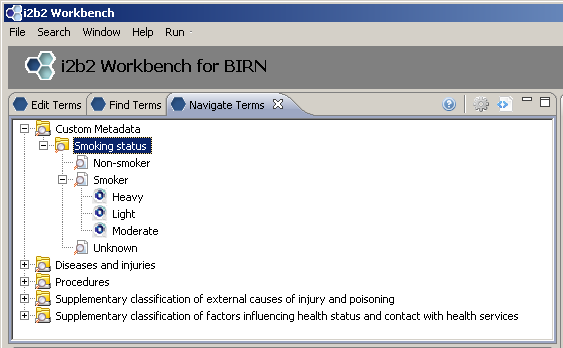
Now we would like to further specify our smokers as “Heavy”, “Moderate” or “Light” smokers. We could create three new terms, or, we can create modifiers for our ‘Smoker’ concept.
Updated ‘Smoker’ concept with modifiers displayed
Using Find Terms Search by Names Tab to Locate Diagnoses

Choose 'Ontology' from the drop-down box, type in a text string ('asthma' in our example), and click on 'Find'.
Using Find Terms Search by Codes Tab to Locate Diagnoses
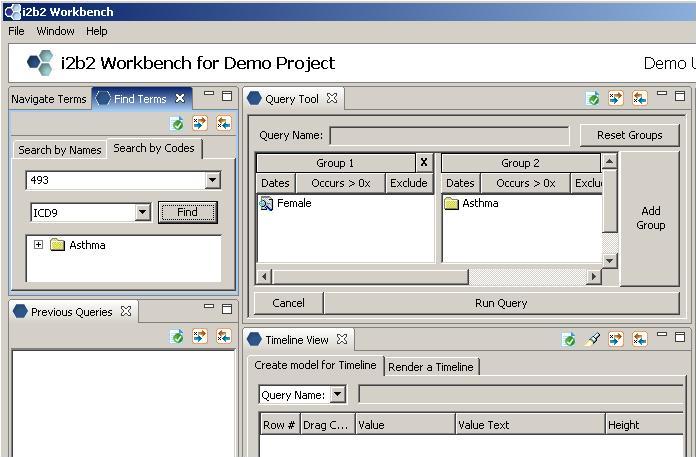
Enter 'ICD-9' from the drop-down box, type in a text string ('493' in our example), and click on 'Find'.
Using Find Terms to Locate Modifier by Name
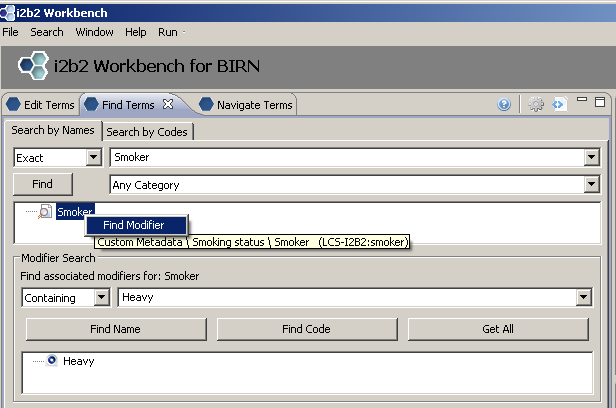
Right click on concept we wish to find modifiers for; Choose ‘Find Modifier’. Enter the name of the modifier we are searching for in text box (‘Heavy’). Select ‘Find Name’
Using Find Terms to Locate Modifier by Code
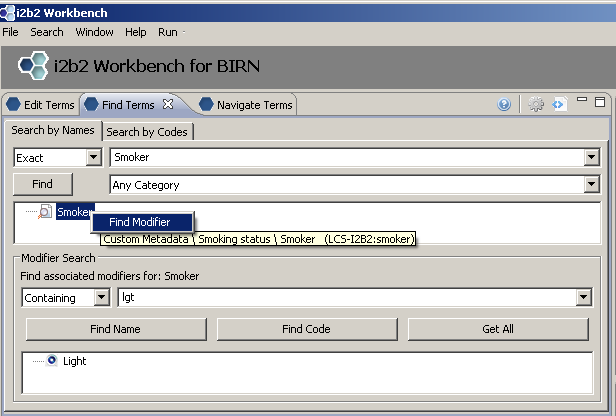
Right click on concept we wish to find modifiers for; Choose ‘Find Modifier’. Enter the code for the modifier we are searching for in text box (‘lgt’). Select ‘Find Code’