DRAFT: This is a work-in-progress!
Totalnum / Patient Counting in i2b2
Counts whisper softly, ETL is wild, Researchers rejoice, Fast counts now arrive, |
|---|
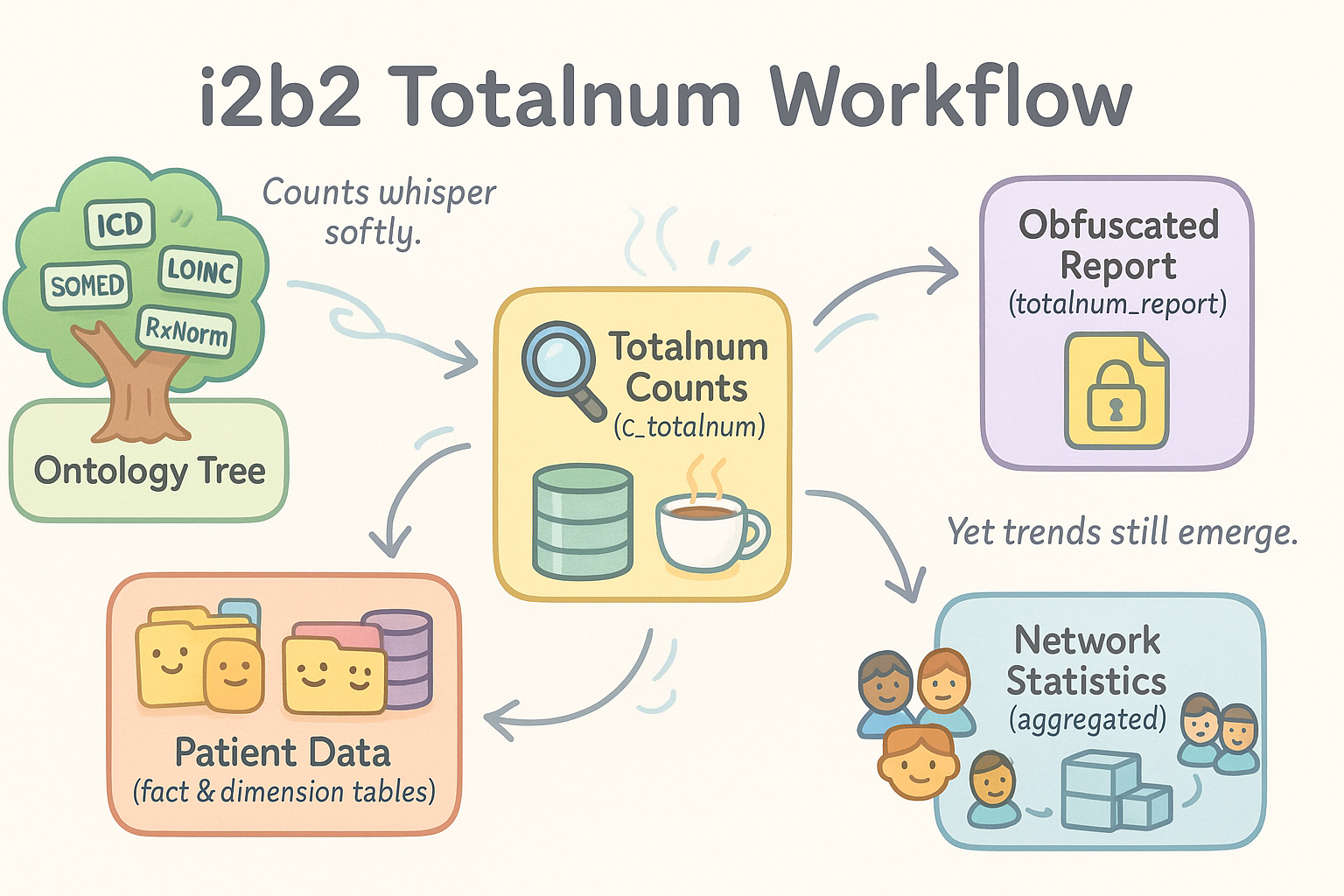
Figure: i2b2 Totalnum Workflow
Ontology terms and patient data feed into the totalnum counting process, producing per-concept patient counts (c_totalnum). These counts are written back into the ontology for query optimization and visualization. Obfuscated versions are stored in totalnum_report, which can then be aggregated across sites to generate privacy-preserving network-level statistics for feasibility studies and data quality assessment.
Overview
The totalnum feature in i2b2 enables sites to calculate and store the number of patients associated with every concept in their ontology. Originally created as a query optimization tool, it has since evolved into a critical component for query development and data quality assessment.
In practice, each i2b2 ontology concept (e.g., a diagnosis code, a lab test, a demographic value) can be associated with the total number of unique patients who have at least one record matching that concept or any of its descendants. These counts can be displayed in the i2b2 web client, helping researchers gauge cohort feasibility before running queries.
Beyond individual sites, federated networks such as ENACT can aggregate these patient counts across institutions. This aggregation supports:
Feasibility studies – researchers can quickly see if a condition is represented across sites.
Data quality checks – missing or unexpected counts can signal mapping errors, ETL problems, or local coding differences.
Ontology usage insights – counts highlight which parts of the ontology are actively populated and which may be underused.
Privacy-preserving comparisons – obfuscation algorithms ensure sensitive patient counts are not directly exposed across sites.
With the introduction of Fast Totalnum in i2b2 v1.8, counts can now be generated 5–10× faster, making it feasible to refresh them regularly as part of ETL workflows.
Technical FAQ
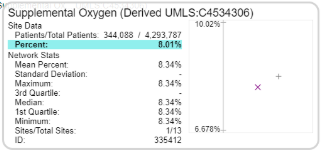
What is | Figure: Counts appear in the ontology tree. Figure: Data Quality Explorer network statistics. |
How do I run Fast Totalnum?
SQL Server (T-SQL)
EXEC FastTotalnumPrep; -- Run once (when ontology changes)
EXEC FastTotalnumCount; -- Run patient counting
EXEC FastTotalnumOutput; -- Write results to ontology + report tables
Oracle (PL/SQL)
BEGIN FastTotalnumPrep; END;
BEGIN FastTotalnumCount; END;
BEGIN FastTotalnumOutput; END;
Postgres
SELECT fasttotalnumprep(); -- Run once (ontology changes)
CALL fasttotalnumcount(); -- Counting step
CALL fasttotalnumoutput(); -- Output results
Can I use totalnum with OMOP data?
Yes. i2b2 v1.8 supports OMOP integration. Use the OMOP-specific totalnum preparation script (e.g., totalnum_fast_prep_OMOP.sql) and configure ontology mappings to OMOP tables such as drug_exposure.
Where are results stored?
Ontology tables (
c_totalnum) → for query optimization and web client display.totalnum_reporttable → obfuscated counts, keyed for cross-site aggregation.

Add Comment