Page History
THIS FEATURE WILL BE AVAILABLE MARCH 2023
Design
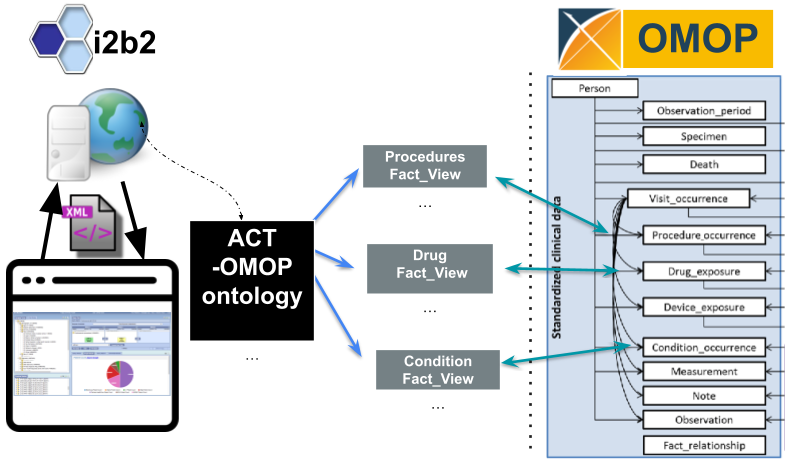
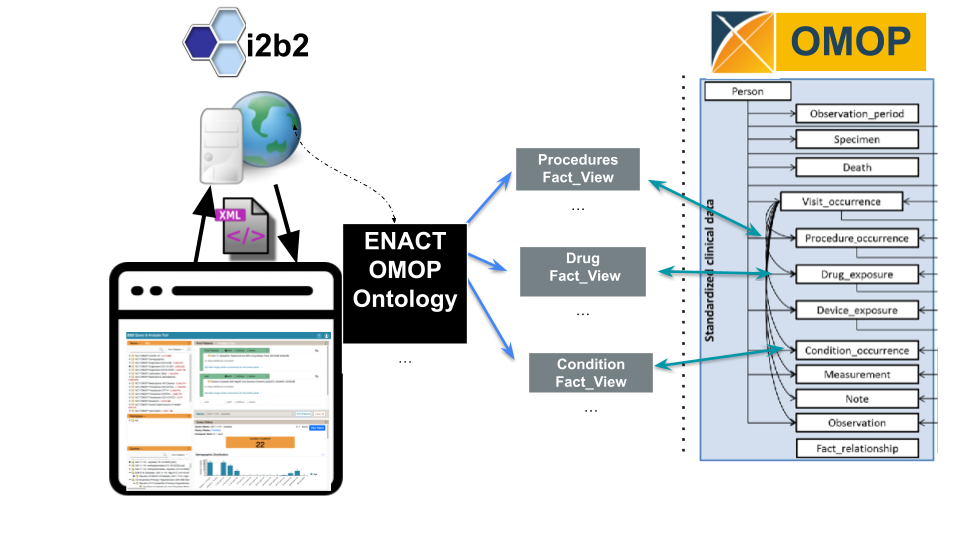
Utilizing i2b2's reusable application programming interface (API)
, the OMOP data model is now supported. The
ENACT-OMOP ontology queries OMOP table views that correspond to the OMOP table structure.
The i2b2 query tool tables and ENACT-OMOP ontology are installed in a he OMOP data and the i2b2 application layer are loaded. With the ontology, it becomes possible to query OMOP tables using the i2b2 interface.
Steps to build ACT-OMOP Ontology
The design document can be found here.
Step 1: Acquire downloads
First, download the necessary components onto a computer that can connect to your target database and from which you can run SQL queries.
- Download i2b2.
- Download and extract the newinstall newi nstall zip package from "Download Binary Distribution" in the top section of https://www.i2b2.org/software
- Determine if you need to install any additional required packages, such as Java JDK or Ant: Chapter 2. Requirements
- If running less than i2b2 1.8, separately download the latest data package from GitHub https://github.com/i2b2/i2b2-data to get the ACT-OMOP ontologies.
- OMOP 5.4 DDL SQL Scripts: https://github.com/OHDSI/CommonDataModel/tree/main/inst/ddl/5.4
- Either clone the repository or download the ETL scripts for your database platform.
Step 2:
CreateInstall i2b2, with ACT-OMOP core tables
and Views (OMOP, i2b2, and ACT-OMOP specific):OMOP:, views, and ontologies.
In 3.4.2- OMOP: If you will be creating a new OMOP database for this installation, then:
- Run the OMOP 5.4 DDL SQL Scripts you downloaded in the last step, using a SQL client.
- It is recommended to run the DDL, indices, and primary key scripts, but NOT the constraints scripts. This can cause problems during ETL.
- This step can be skipped if you already have OMOP on your target database.
- Run the OMOP 5.4 DDL SQL Scripts you downloaded in the last step, using a SQL client.
- If you have not installed i2b2 previously (i.e. you are not setting this up as an i2b2 Project in a larger installation):
- i2b2: You will be installing i2b2 as explained in the installation guide. Follow the chapters in the installation guide, but see the notes in the next bullet on Chapter 3.
- Chapter 3. Data Installation requires special handling. All the steps in the installation guide should be followed except for the following changes in the CRC and Metadata sections::
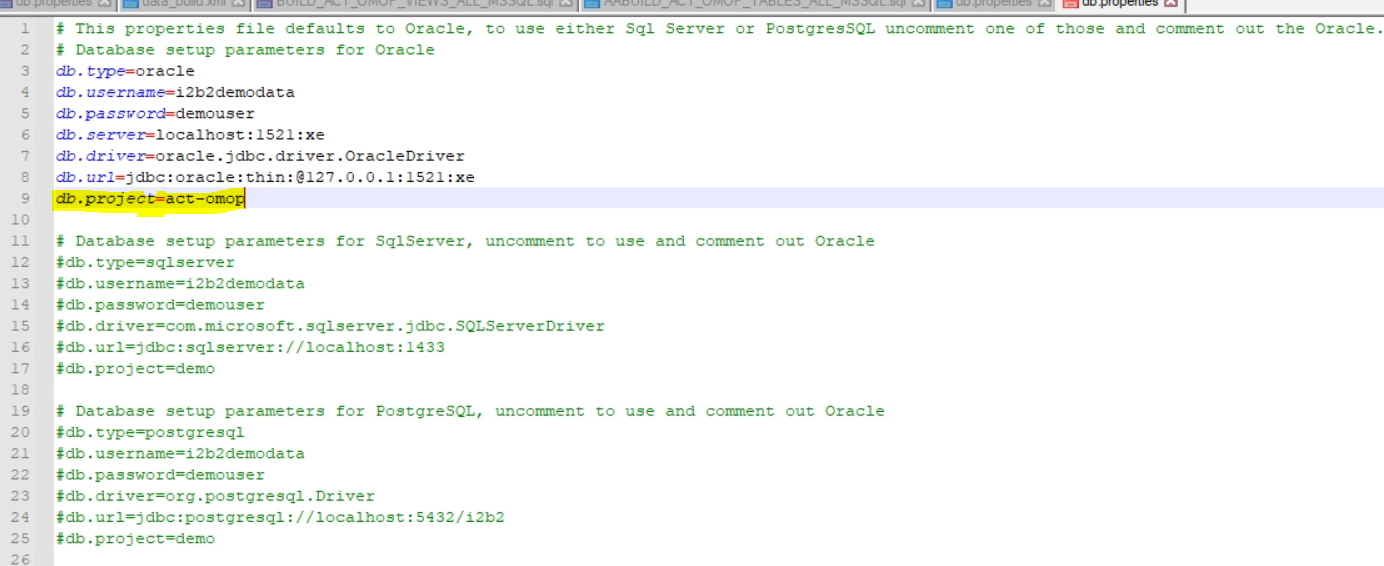
- You need to select the act-omop project for Metadata and CRC. Specifically, in 3.4.2 Set Database Properties (CRC) and 3.7.2 Set Database Properties (Metadata), edit the CRC db properties file to set db.project parameter to act-omop.
- Instead of CRC install step 3.4.
- 4, drop the patient_dimension and visit_dimension tables from the database. For convenient reference, the correct steps 3.4.3-3.4.5 are summarized here:
ant -f data_build.xml create_crcdata_tables_release_<version no>
FROM THE DATABASE: DROP PATIENT_DIMENSION; DROP VISIT_DIMENSION;
ant -f data_build.xml create_procedures_release_<version no>
ant -f data_build.xml db_demodata_load_data
- 4, drop the patient_dimension and visit_dimension tables from the database. For convenient reference, the correct steps 3.4.3-3.4.5 are summarized here:
The final ant script will create act-omop Views, which point to the OMOP tables, and loads the concept_dimension table with act-omop concepts data
| Info |
|---|
You will need to drop PATIENT_DIMENSION and VISIT_DIMENSION before you load any data. |
Step 3: ACT-OMOP v4 ontology data load process
. The first two steps create the standard CRC tables and procedures.
In 3.7.4, only load the regular data, not the identified data. Run the ant command:
ant -f data_build.xml db_metadata_load_data
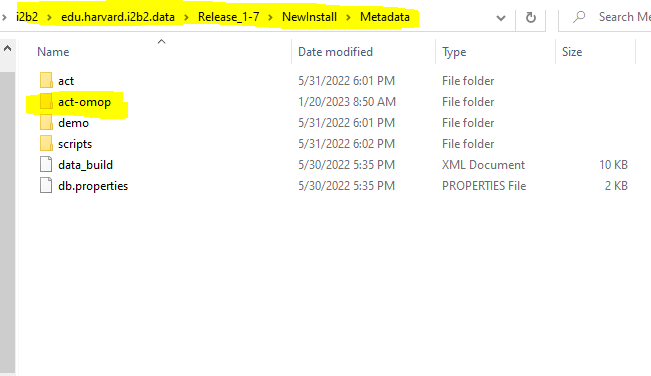
- This will execute the SQL scripts from the edu.harvard.i2b2.data\Release_
- <version no>\NewInstall\Metadata\act-omop\scripts\<db type> folder and:
- Creates ACT-OMOP v4.1 Ontology metadata tables with indexes.
- Loads the act-omop metadata
- <version no>\NewInstall\Metadata\act-omop\scripts\<db type> folder and:
- Make sure i2b2 is configured for multiple fact tables by changing the appropriate entry in the database.
| Info |
|---|
Be sure to drop/rename PATIENT_DIMENSION and VISIT_DIMENSION tables before you run the db_demodata_load_data ant target. |
| Info |
|---|
The Lab metadata tables should have data in the C_METADATAXML column in order for the lab values to be queriable from UI If the C_METADATAXML is missing from the source ontology Lab metadata file, you can load the values from the standard i2b2-ACT Lab metadata table. |
Step 4: Create Project
Finally| Info |
|---|
The CPT4 ontology table is not included with i2b2 due to AMA restrictions on redistribution of CPT code information. Contact the ACT team to get a copy if your institution is an AMA member. |
Step 3: Advanced Users - Create Project
If an OMOP project will coexist with an i2b2 project, set up an i2b2 OMOP project pointing to the i2b2-OMOP database that you just configured.
6.6.0. [DRAFT] Guide to creating a new project with the admin tool.
Verify the Ontology is displayed and queriable from the webclient
| Info |
|---|
The concept_Dimension is populated from metadata tables. Verify create_omop_act_concept_dimension.sql to ensure it has complete metadata tables information before running the script Example: the ACT/OMOPVital_Signs information should be in the script to load the ACT/OMOPVital_Signs concept data |
Notes
Both the ACT-OMOP Ontology tables, views and data is available under edu.harvard.i2b2.data\Release_1-7<version no>\NewInstall\Metadata\act-omop folder.
The scripts are in this folder:
These are the ACT-OMOP ontology tables.
ACT_ICD10CM_DX_V4V41_OMOP ACT_ICD10PCS_PX_V4V41_OMOP ACT_ICD9CM_DX_V4V41_OMOP ACT_ICD9CM_PX_V4V41_OMOP ACT_CPT4_PX_V4_OMOPACT_HCPCS_PX_V4V41_OMOP ACT_MED_ALPHA_V4V41_OMOP ACT_MED_VA_V4V41_OMOP ACT_MEDLOINC_VALAB_V4PROV_V41_OMOP ACT_LOINC_LAB_PROVV41_V4_OMOP ACT_SDOH_V4V41_OMOP ACT_VITAL_SIGNS_V4V41_OMOP ACTACT_VISIT_DETAILS_V4V41_OMOP ACT_COVID_V4V41_OMOP ACT_DEM_V4V41_OMOP ACT_ICD10_ICD9_DX_V4V41_OMOP ACT_RESEARCH_V41_OMOP ACT_VAX_V41_OMOP ACT_ZIPCODE_V41_OMOP |
These are the ACT-OMOP views:
EMPTY_VIEW; * this is actually an empty table CONDITION_VIEW; DRUGDRUG_VIEW; MEASUREMENTMEASUREMENT_VIEW; OBSERVATIONOBSERVATION_VIEW; PROCEDURE_VIEW; COVID_LAB_VIEW; ALL PATIENT_SOURCE_CONCEPTSDIMENSION; PATIENTVISIT_DIMENSION; VISIT DEVICE_DIMENSIONVIEW; DEVICE CONDITION_NS_VIEW; DRUG_NS_VIEW; MEASUREMENT_NS_VIEW; OBSERVATION_NS_VIEW; PROCEDURE_NS_VIEW; DEVICE_NS_VIEW; VISIT_NS_VIEW; |
- ETL OMOP data
- This is a site-specific process, but OHDSI's WhiteRabbit tool can help: https://github.com/OHDSI/WhiteRabbit