Page History
...
Patient Data Object (PDO)
Document Version: | 1.4.1 |
I2b2 Software Release: | 1.4 |
Table of Contents
i2b2 XML Schema Definitions Definitions
| Anchor | ||||
|---|---|---|---|---|
|
...
The Informatics for Integrating Biology and the Bedside (i2b2) is one of the sponsored initiatives of the NIH Roadmap National Centers for Biomedical Computing (http://www.bisti.nigh.gov/ncbc). One of the goals of i2b2 is to provide clinical investigators broadly with the software tools necessary to collect and manage project-related clinical research data in the genomics age as a cohesive entity - – a software suite to construct and manage the modern clinical research chart. The i2b2 hive is a set of cells or modules that have a common messaging protocol that allow the cells to interact using web services and XML messages.
...
All cells in the i2b2 hive must communicate using standard i2b2 XML messages. This message specifies certain properties that are common to cells and essential to the administration tasks associated with sending, receiving and processing messages.
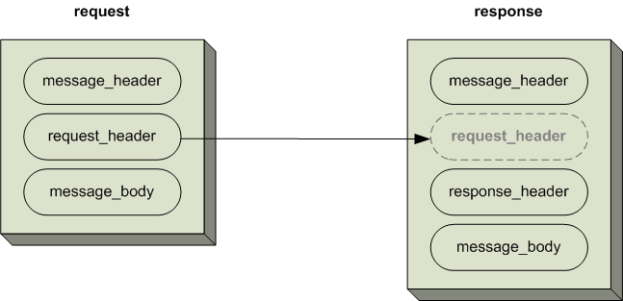
All requests are sent using a <request> tag and responses are returned using a <response> tag. The same <message_header> tag is used for both. The <request_header> is used for requests but may optionally be echoed back in the response. The response must include a <response_header>.
The XSD specification of the i2b2 message permits individual cells to add cell-specific XML in the <message_body> tag. This cell-specific XML need not extend the i2b2 message schema since the i2b2 schema will allow insertion of tags from any namespace into the <message_body> tag.
The following image illustrates the basic top-level elements contained within the request and response messages.
...
<repository:patient_data xmlns:repository="http://i2b2.mgh.harvard.edu/repository_cell:
xsi:schemaLocation="http://i2b2.mgh.harvard.edu/repository_cell/patient_data.xsd">
<event_set>
<event update_date="2006-05-04T18:13:51.0Z"
download_date="2006-05-04T18:13:51.0Z"
import_date="2006-05-04T18:13:51.0Z"
sourcesystem_cd="sourcesystem_cd0"
upload_id="upload_id0">
<event_id source="HIVE">event_id0</event_id>
<patient_id source="HIVE">patient_id0</patient_id>
<param column="inout_cd" name="in vs. outpatient code"></param>
<param column="location_cd" name="location code"></param>
<param column="location_path" name="location hierarchy"></param>
<param column="active_status_cd" name="date interpretation code"></param>
<start_date>2006-05-04T18:13:51.0Z</start_date>
<end_date>2006-05-04T18:13:51.0Z</end_date>
<event_blob></event_blob>
</event>
< </event_set>
<concept_set>
<concept update_date="2006-05-04T18:13:51.0Z"
download_date="2006-05-04T18:13:51.0Z"
import_date="2006-05-04T18:13:51.0Z"
sourcesystem_cd="sourcesystem_cd3"
upload_id="upload_id3">
<concept_path>concept_path0</concept_path>
<concept_cd>concept_cd0</concept_cd>
<name_char>name_char0</name_char>
<concept_blob></concept_blob>
</concept>
</concept_set>
<observer_set>
<observer update_date="2006-05-04T18:13:51.0Z"
download_date="2006-05-04T18:13:51.0Z"
import_date="2006-05-04T18:13:51.0Z"
sourcesystem_cd="sourcesystem_cd6" upload_id="upload_id6">
<observer_path>observer_path0</observer_path>
<observer_cd>observer_cd0</observer_cd>
<name_char>name_char3</name_char>
<observer_blob></observer_blob>
</observer>
</observer_set>
<pid_set>
<pid>
<patient_id status="status0" update_date="2006-05-04T18:13:51.0Z"
download_date="2006-05-04T18:13:51.0Z"
import_date="2006-05-04T18:13:51.0Z"
sourcesystem_cd="sourcesystem_cd9"
upload_id="upload_id9" source="HIVE">patient_id3</patient_id>
<patient_map_id source="HIVE" status="status1"
update_date="2006-05-04T18:13:51.0Z"
download_date="2006-05-04T18:13:51.0Z"
import_date="2006-05-04T18:13:51.0Z"
sourcesystem_cd="sourcesystem_cd10"
upload_id="upload_id10">patient_map_id0</patient_map_id>
</pid>
< </pid_set>
<eid_set>
<eid>
<event_id source="HIVE" patient_id="patient_id6" patient_id_source="HIVE" status="status6" update_date="2006-05-04T18:13:51.0Z"
download_date="2006-05-04T18:13:51.0Z"
import_date="2006-05-04T18:13:51.0Z"
sourcesystem_cd="sourcesystem_cd15"
upload_id="upload_id15">event_id3</event_id>
<event_map_id source="HIVE" patient_id="patient_id7"
patient_id_source="HIVE" status="status7"
update_date="2006-05-04T18:13:51.0Z"
download_date="2006-05-04T18:13:51.0Z"
import_date="2006-05-04T18:13:51.0Z"
sourcesystem_cd="sourcesystem_cd16"
upload_id="upload_id16">event_map_id0</event_map_id>
< </eid>
< </eid_set>
<patient_set>
<patient update_date="2006-05-04T18:13:51.0Z"
download_date="2006-05-04T18:13:51.0Z"
import_date="2006-05-04T18:13:51.0Z"
sourcesystem_cd="sourcesystem_cd21"
upload_id="upload_id21">
<patient_id source="HIVE">patient_id12</patient_id>
<param column="vital_status_cd" name="date interpretation code">param3</param>
<param column="birth_date" name="birthdate">param3</param>
<param column="death_date" name="date of death">param3</param>
<param column="sex_cd" name="gender">param3</param>
<param column="age_in_years_num" name="age">param3</param>
<param column="language_cd" name="language">param3</param>
<param column="race_cd" name="race">param3</param>
<param column="religion_cd" name="religion">param3</param>
<param column="marital_status_cd" name="marital status">param3</param>
<param column="statecityzip_path_char" name="zip code hierarchy">param3</param>
<patient_blob></patient_blob>
</patient>
< </patient_set>
<observation_set panel_name="panel_name0">
<observation update_date="2006-05-04T18:13:51.0Z"
download_date="2006-05-04T18:13:51.0Z"
import_date="2006-05-04T18:13:51.0Z"
sourcesystem_cd="sourcesystem_cd24"
upload_id="upload_id24">
<event_id source="HIVE">event_id6</event_id>
<patient_id source="HIVE">patient_id15</patient_id>
<concept_cd name="name6">concept_cd3</concept_cd>
<observer_cd soruce="soruce0">observer_cd3</observer_cd>
<start_date>2006-05-04T18:13:51.0Z</start_date>
<modifier_cd name="name7">modifier_cd0</modifier_cd>
<valuetype_cd>valuetype_cd0</valuetype_cd>
<tval_char>tval_char0</tval_char>
<nval_num units="units0">3.141592653589</nval_num>
<valueflag_cd name="name8">valueflag_cd0</valueflag_cd>
<quantity_num>3.141592653589</quantity_num>
<units_cd>units_cd0</units_cd>
<end_date>2006-05-04T18:13:51.0Z</end_date>
<location_cd name="name9">location_cd0</location_cd>
<confidence_num>3.141592653589</confidence_num>
<observation_blob></observation_blob>
</observation>
</observation_set>
</repository:patient_data>
...
| Anchor | ||||
|---|---|---|---|---|
|
| Anchor | ||||
|---|---|---|---|---|
|
Object |
| Explanation |
concept_set | Data from the concept_dimension table |
|
concept | One row of data from the concept_dimension table. |
|
concept_path | A path that represents the hierarchical specification of the concept |
|
concept_cd | A unique code that represents a concept. |
|
name_char | A string name that represents this concept, idea or person. |
|
concept_blob | XML data that includes partially structured and unstructured data about a concept. |
|
| Anchor | ||||
|---|---|---|---|---|
|
| Anchor | ||||
|---|---|---|---|---|
|
Object |
| Explanation |
observer_set | Data from the provider_dimension table |
|
observer | One row of data from the provider_dimension table. |
|
observer_path | A path that represents the hierarchical specification of the observer |
|
observer_cd | A unique code that represents a observer. |
|
name_char | A string name that represents this observer, could be person or machine. |
|
observer_blob | XML data that includes partially structured and unstructured data about an observer. |
|
| Anchor | ||||
|---|---|---|---|---|
|
| Anchor | ||||
|---|---|---|---|---|
|
Object |
| Explanation |
pid_set | Data from the patient_mapping table |
|
pid | One set of mappings on a single patient_num |
|
patient_id | A choice between Patient_Num, (if source is HIVE) or Patient_Id if another source. A source with "_e" at the end is encrypted. |
|
patient_map_id | A patient_id that should have the same patient_num as the patient_id in this pid. |
|
| Anchor | ||||
|---|---|---|---|---|
|
| Anchor | ||||
|---|---|---|---|---|
|
Object |
| Explanation |
eid_set | Data from the encounter_mapping table |
|
eid | One set of mappings on a single visit_num |
|
event_id | A choice between visit_num, (if source is HIVE) or Visit_Id if another source. A source with "_e" at the end is encrypted. |
|
event_map_id | A visit_id that should have the same patient_num as the visit_id in this eid. |
|
| Anchor | ||||
|---|---|---|---|---|
|
| Anchor | ||||
|---|---|---|---|---|
|
Object |
| Explanation |
observation_set | Data from the observation_fact table. |
|
Observation | One row of data from the observation_fact table. |
|
event_id | A choice between encounter_num (if source is HIVE) or encounter_id if an other source. A source with "_e" at the end is encrypted. |
|
patient_id | A choice between patient_num, (if source is HIVE) or patient_id if another source. A source with "_e" at the end is encrypted. |
|
concept_cd | A unique code that represents a concept. |
|
observer_cd | An ID that represents the provider, which could be a physician or a machine such as an MRI machine. |
|
start_date | The date that the observation was made, or that the observation started. If the data is derived or calculated from another observation (like a report) then the start date is the same as the observation it was derived or calculated from. |
|
modifier_cd | hierarchical derivations of a common observation |
|
ValType_Cd | A code representing whether a value is stored in the TVal column, NVal column, or observation_blob column. |
|
TVal_Char | A text value. |
|
NVal_Num | A numerical value. |
|
ValueFlag_Cd | A code that represents the type of value present in the NVal_Num, the TVal_Char or observation_blob column |
|
Quantity_Num | The number of observations represented by this fact. |
|
Units_Cd | A textual description of the units associated with a value. |
|
End_Date | The date that the observation ended. If the data is derived or calculated from another observation (like a report) then the end_date is the same as the observation it was derived or calculated from. |
|
Location_Cd | A code representing the hospital associated with this visit. |
|
Confidence_Num | A code or number representing the confidence in the accuracy of the data. |
|
Observation_Blob | XML data that includes partially structured and unstructured data about an observation. |
|
| Anchor | ||||
|---|---|---|---|---|
|
| Anchor | ||||
|---|---|---|---|---|
|
Object |
| Explanation |
patient_set | data from the visit_dimension table |
|
patient | One row of data from the visit_dimension table. |
|
patient_id | A choice between patient_num, (if source is HIVE) or patient_id if another source. A source with "_e" at the end is encrypted. |
|
start_date | The date-time that the patient was born. |
|
end_date | The date-time that the patient died. |
|
vital_status_cd | A code to represent the meaning of the date fields above. |
|
param | Name value field |
|
patient_blob | XML data that includes partially structured data about a patient |
|
| Anchor | ||||
|---|---|---|---|---|
|
| Anchor | ||||
|---|---|---|---|---|
|
...
Object | Explanation |
Update_Date | The date the data was last updated according to the source system from which the data was obtained. If the source system does not supply this data, it defaults to the download_date. |
Download_Date | The date that the data was obtained from the source system. If the data is derived or calculated from other data, then the download_date is the date of the calculation. |
Import_Date | The date the data is placed into the table of the data mart. |
SourceSystem_Cd | A code representing the source system that provided the data. |
Upload_Id | Tracking number assigned to any file uploaded. |
Document Version: | 1.4.1 |
I2b2 Software Release: | 1.4 |