Page History
...
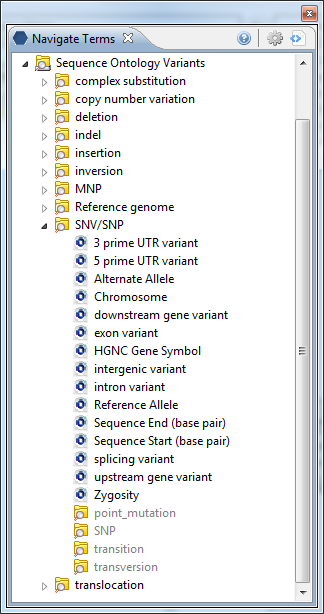
A subset of the Sequence Ontology (http://www.sequenceontology.org) describing variants was repurposed for this project. The ontology organizes a set of concepts describing the structural change of the variant, such as a SNP/SNV (Single nucleotide polymorphism/Single nucleotide variant), or the insertion or the deletion of bases. Attributes of the variant are expressed as "modifiers" for that variant. In their simplest form shown below they may represent information relating to the location of the variant: start and end location of the variant within a chromosome; the gene in which the variation is thought to occur; or the type of variant (exonic, intronic, intergenic). The tree shown here appears on the http;//www.i2b2.org/webclient demo site.
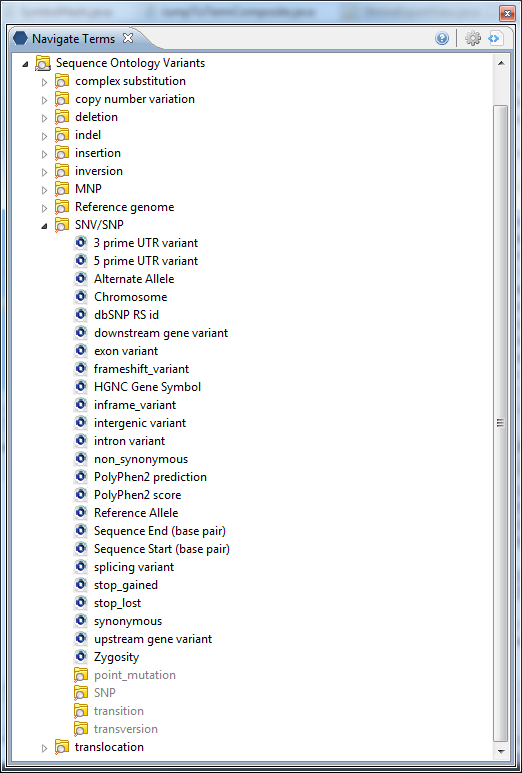
A More Complicated i2b2 Sequence Ontology
Many tools exist to annotate NGS output vcf data files.With these annotations come opportunities to represent even more modifiers on the Navigation Tree. For example we can expand the tree to add exon-centric features such as dbSNP numbers,non/synonymous variant classification, PolyPhen scores, and functional attributes such as stop_loss or stop_gain. In essence, you can grow or shrink the ontology (and the observation fact data collected) as your system requirements dictate.